pip install git+https://github.com/wayneweiqiang/GaMMA.gitThe implementation is based on the Gaussian mixture models in scikit-learn
- Zhu, Weiqiang et al. "Earthquake Phase Association using a Bayesian Gaussian Mixture Model." (2021)
- Zhu, Weiqiang, and Gregory C. Beroza. "PhaseNet: A Deep-Neural-Network-Based Seismic Arrival Time Picking Method." arXiv preprint arXiv:1803.03211 (2018).
- Hyperparameters:
- use_amplitude (default = True): If using amplitude information.
- vel (default = {"p": 6.0, "s": 6.0 / 1.75}): velocity for P and S phases.
- use_dbscan: If using dbscan to cut a long sequence of picks into segments. Using DBSCAN can significantly speed up associaiton using small windows.
- dbscan_eps (default = 10.0s): The maximum time between two picks for one to be considered as a neighbor of the other. See details in DBSCAN
- dbscan_min_samples (default = 3): The number of samples in a neighborhood for a point to be considered as a core point. See details in DBSCAN
- oversampling_factor (default = 10): The initial number of clusters is determined by (Number of picks)/(Number of stations)/(Inital points) * (oversampling factor).
- initial_points (default=[1,1,1] for (x, y, z) directions): Initial earthquake locations (cluster centers). For a large area over 10 degrees, more initial points are helpful, such as [2,2,1].
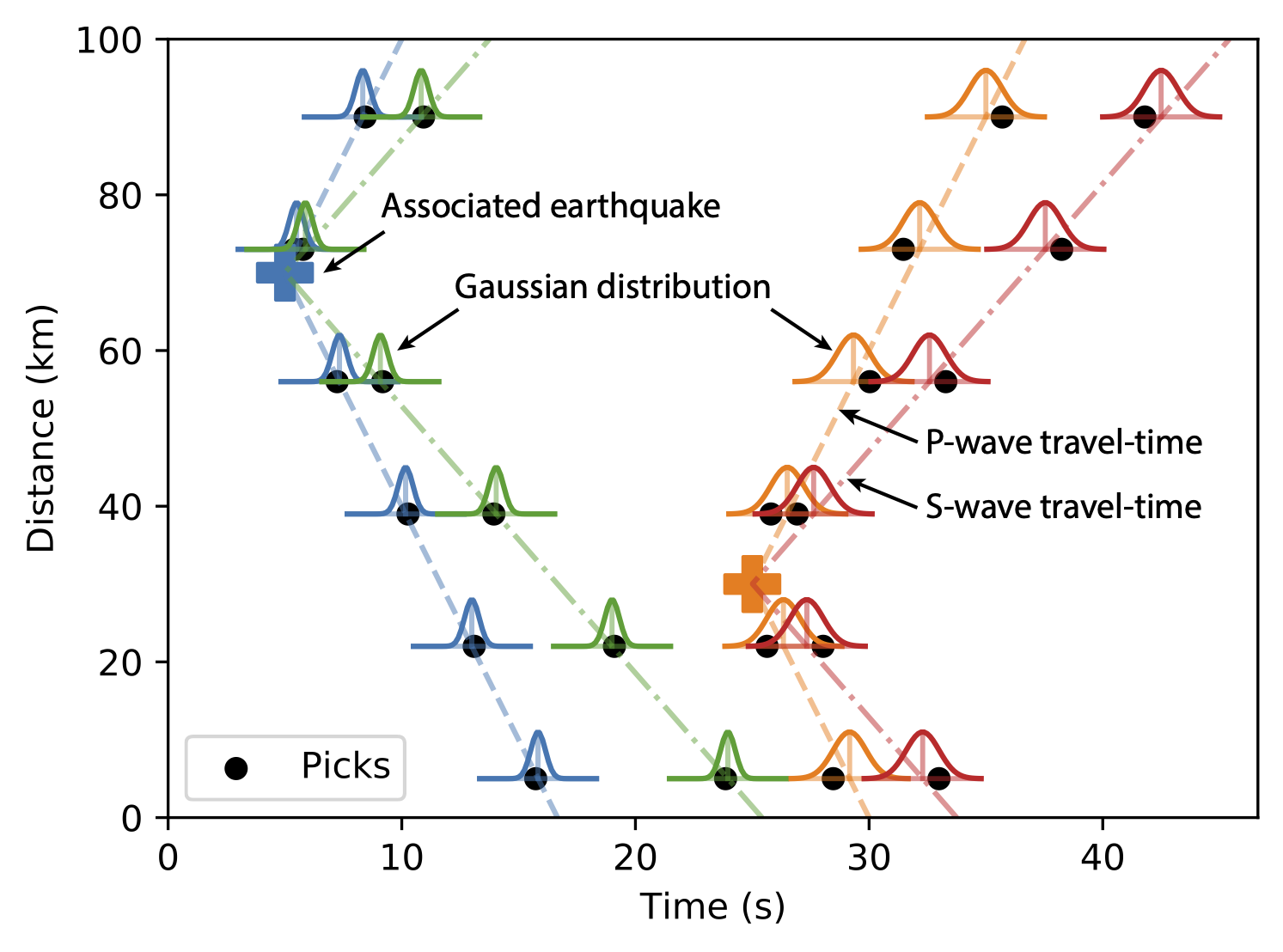
- covariance_prior (default = (5, 5)): covariance prior of time and amplitude residuals. Because current code only uses an uniform velocity model, a large covariance prior can be used to avoid splitting one event into multiple events.
- Filtering low quality association
- min_picks_per_eq: Minimum picks for associated earthquakes. We can also specify minimum P or S picks:
- min_p_picks_per_eq: Minimum P-picks for associated earthquakes.
- min_s_picks_per_eq: Minimum S-picks for associated earthquakes.
- max_sigma11: Max phase time residual (s)
- max_sigma22: Max phase amplitude residual (in log scale)
- max_sigma12: Max covariance term. (Usually not used)
Note the association speed is controlled by dbscan_eps and oversampling_factor. Larger values are preferred, but at the expense of a slower association speed.
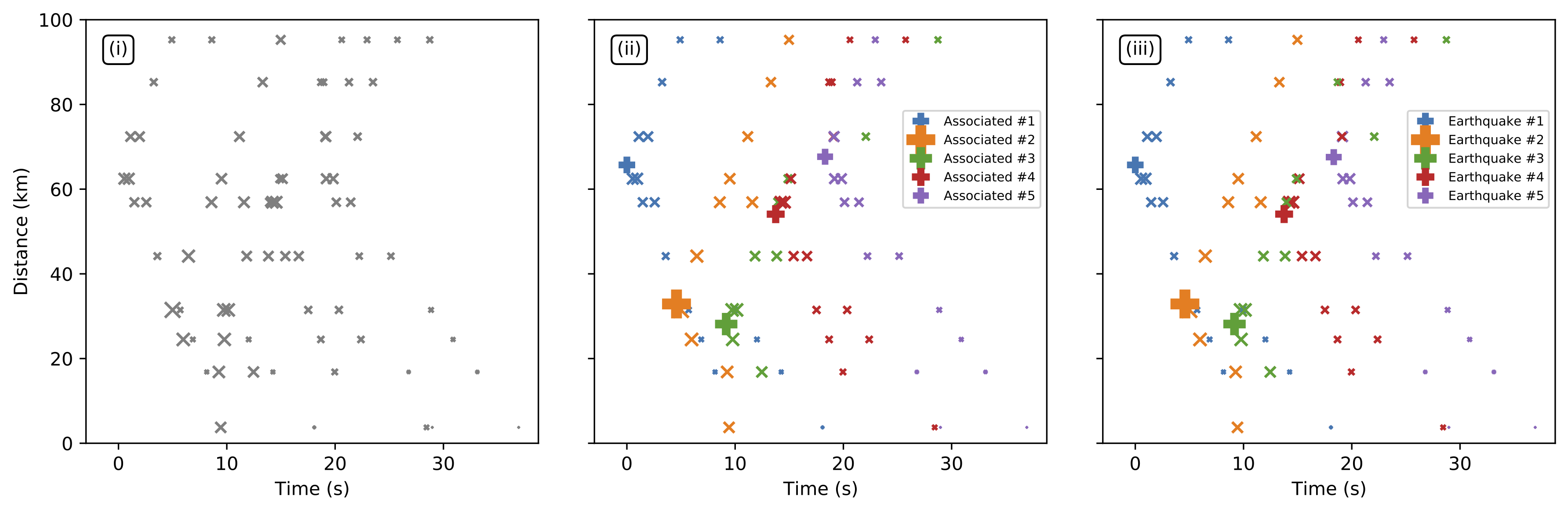
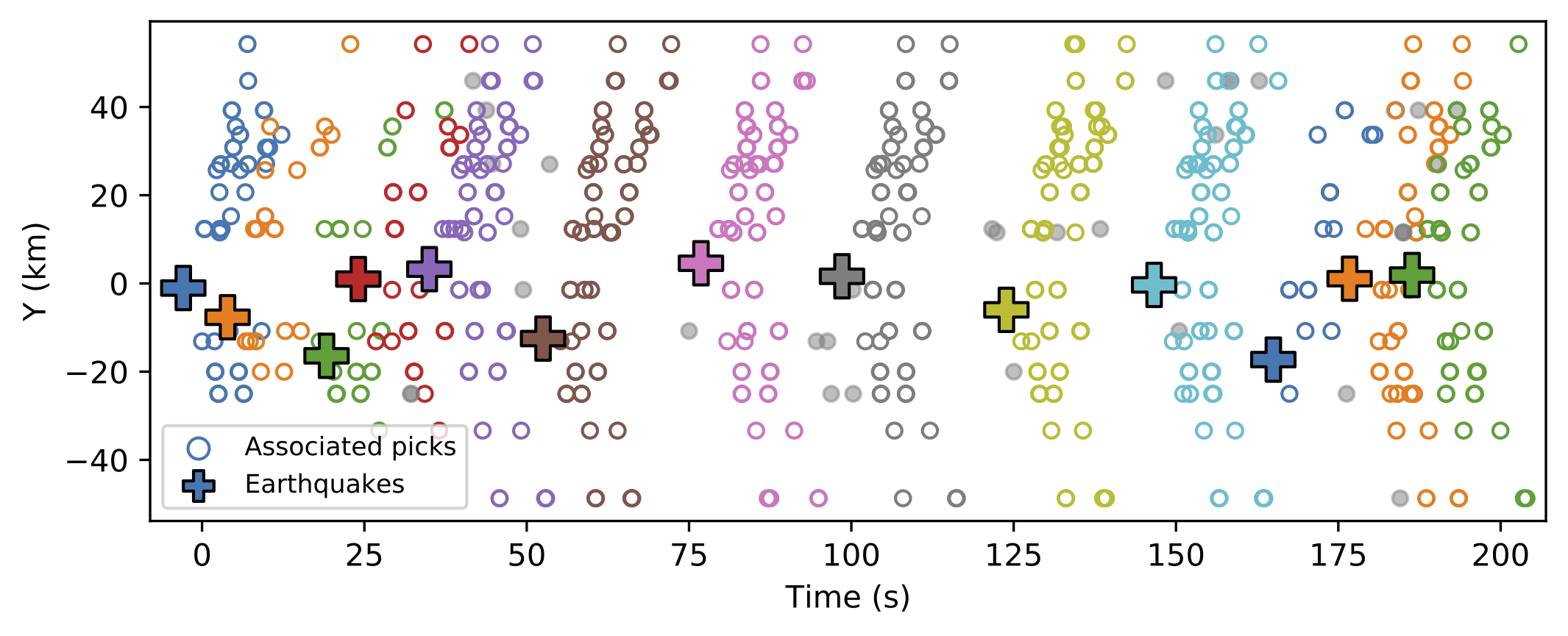
- Synthetic Example
- Example using PhaseNet picks
See details in the notebook: example_phasenet.ipynb
- Example using Seisbench
See details in the notebook: example_seisbench.ipynb
More examples can be found in the earthquake detection workflow -- QuakeFlow