tolstikhin / wae Goto Github PK
View Code? Open in Web Editor NEWWasserstein Auto-Encoders
License: BSD 3-Clause "New" or "Revised" License
Wasserstein Auto-Encoders
License: BSD 3-Clause "New" or "Revised" License
Problem is the following When I run the run.py .
tensorflow.python.framework.errors_impl.UnknownError:
NewRandomAccessFile failed to Create/Open: ./mnist\train-images-idx3-ubyte
Input/output error
Hi,
Thanks for your shared codes and I learned so much out of it!
I'm going to modify your wae implementation for 1D sequential data (protein sequences reconstruction), However, your codes are hugely image specific.
Can you help me with that?!
Hi,
Thank you so much for this nice implementation. However i have two questions about the objective function of wae:
best wishes
zhangyiyang
Hi,
I am little bit confused after reading your paper. Please correct me if I misunderstood.
In your paper, you show difference between VAE and WAE in terms of distribution matching objective
However, I wonder why you implemented WAE that matches Q(z|x) to P(z) with GAN and MMD.
(ex. Discriminator still discriminates z tilda from Q(z|x) and z from P(z)
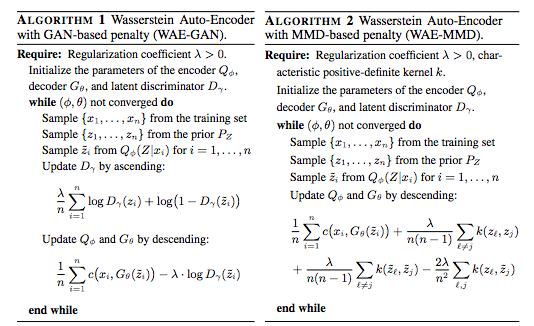
In order to match Q(z) to P(z), don't you have to calculate distance between Q(z) and P(z), in which Q(z) is obtained by marginalizing Q(z|x) with P(x)?
But you are averaging distance between Q(z|x) and P(z) with multiple x.
Hi,
I am investigating how to implement WAE for a fully convolutional encoder and decoder such that there is no fully connected layers being used. Assuming that I am working with 1D data, I have a latent code (i.e. output of the bottleneck) with dimensionality [batch_size X 1024 channels X 8 samples_per_channel].
I have used the implementation of imq_kernel shown here which is similar to your implementation . However, this is only working with 2D data (no channel dimension).
My question: is it OK to sum every tensor along the channel dimension (which will lead to matrix of size [batch_size X number_of_samples] then continue as usual ? or there is no way from using fully connected layer to flatten the tensor before mmd calculation ?
When I have used that approach, I got very unstable values for the mmd_loss, the values are fluctuating between positive and negative and there is no monotonic decrease !
Finally: in case of using fully conv layers like what I have described, does the latent code represent a samples from distribution Q(z|x) that should be compared w.r.t Gaussian noise of mean 0 and covariance I ? Or the latent in all cases shall be a vector of means and matrix of co-variance that should be fitted onto Gaussian (which is ur case when using FC layers)?! Does the situation (of fitting mean and covar onto Gaussian) happen also in case of WAE-GAN ?
Sorry for prolonging and many thanks in advance
Hi,
Thanks for this interesting paper and implementation :)
My question: why do we still need discriminator network for the WAE-MMD approach ? As stated in algorithm 2 of the paper ?
Best REgards
This error appears when using the DCGAN as encoder / decoder.
Exactly in this line :
h0 = tf.reshape(h0, [-1, height, width, num_units])
and height, width are floats.
I don't get the formula to get the value of heights and widths.
Can you explain it more please ?
Thank you in advance :)
Hello,
Correct me if I'm wrong but my understanding is that the simplified expression of the Wasserstein distance, obtained in theorem 1 relies heavily on the hypothesis that the latent codes distribution matches exactly the prior.
But with the necessary relaxation on this constraint, the hypothesis doesn't hold. Do you have any sense of what is happening when the constraint is "violated too much" (e.g. lambda is too small...)?
I haven't had time to run an empirical study and can't wrap my head around what it implies "theoretically".
Any insight to share?
Also, in your implementation, I notice there is an "implicit" model of noise for the encoder. I understand that the noise is parameterized by a neural network that is learnt along in a training of the WAE but can you give a bit more of an insight about it? I can't find any reference to it in the WAE paper or any of the follow-ups I know. Any pointer?
Thanks.
Dear Tolstikhin
Thanks for your solid work, but I want to consult you about the MMD loss function during my specific training: I found that MMD loss function is negative when I was training other datasets except for MNIST and CelebA. Why did that circumstance happen and what did that mean? How should I tackle the negative loss function?
Thanks for sharing a code with your amazing paper! I really enjoyed reading it.
Anyway, I am interested in extending your work in other direction, and I come up with a question on MMD part. I was able to understand the overall concept, but not sure on this multi-scale part.
Line 294 in 068a257
Are you just trying multiple kernels to get a better estimate of MMD?
It would be also very nice of you to recommend some readings to get a better understanding of MMDS.
I am currently using TTUR implementation to compute FID scores where we need to pass 2 things i.e <path_to_generated_samples> and <path_to_original_images> or <path_to_pre-computed_statistics_file>.
Now I am confused for the value of FID corresponding to True data(reference to celebA) i.e how do we evaluate it?
According to my understanding, the FID for true data is computed in following steps :
So is this same process you followed for computing FID ?
So if above is correct then further speculating I can compute FID by generating samples from my generator which construct images of 64x64 -- say G1 directory. Then to compare FID for this model we just need to run "fid.py <path_to_G1> <path_to_S1/S2>" ?
PS : I did not find much resources explaining the end to end procedure for computing FID, so asked here. By the way I really enjoyed your paper reading. It's very well written, learnt a lots of maths !!
Regards,
Prateek
Hello Sir,
I saw that in the reconstruction_loss function (wae.py) you multiply L2, L2_2 and L1 losses with different kind of values, but I haven't read anything about this in the paper.
Can you please explain?
Thank you,
Adi Zholkover
Hi,
Thank you so much for this nice implementation. I am trying to readapt it to my input dataset.
However, I didn't get this parameter which is num_units or num_filters. Do you mean by that number of neurons ? According to what you chose 1024 ?
It is not clear for me, because I assume that the encoder-decoder architecture use not the same number of units for each layer. So for the encoder , the number of units decreases from the dimension of the image which is 28*28 to the latent dimension. For the decoder, it is the opposite process.
I assumed this because Wasserstein autoencoders are quite similar to the variational autoencoder but without the discriminator.
Can I know why you assume that all the layers have the same number of units ?
Thank you in advance.
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.