Comments (31)
Well... in my test, the MNE version worked even better!
import mne
from pathlib import Path
import numpy as np
from mnelab.io.writers import write_set
import matlab.engine
from mne.externals.pymatreader import read_mat
from pyprep.prep_pipeline import PrepPipeline
eeg_file_path = "/media/christian/ElementsSE/lewis/EEG/PaDa.bdf"
raw = mne.io.read_raw_bdf(eeg_file_path, verbose=False)
raw.load_data(verbose=False)
raw.drop_channels(["EXG1", "EXG2", "EXG3", "EXG4", "EXG5", "EXG6", "EXG7", "EXG8", "Status"])
channels = read_mat(str(Path('./connectivityanalyses/MatlabPipeline/channel.mat').absolute()))
ch_pos = {name: loc[[1, 0, 2]] for name, loc in zip(channels["Channel"]["Name"], channels["Channel"]["Loc"])
if name not in ["EXG1", "EXG2", "EXG3", "EXG4", "Status"]}
montage = mne.channels.make_dig_montage(ch_pos=ch_pos,
nasion=None, # fiducials[subject][1]["NAS"][[1, 0, 2]]/1000.0,
lpa=None, # fiducials[subject][1]["LPA"][[1, 0, 2]]/1000.0,
rpa=None) # fiducials[subject][1]["RPA"][[1, 0, 2]]/1000.0)
raw.set_montage(montage)
raw = raw.crop(tmax=120.0)
write_set("/home/christian/Desktop/test_data/test_raw.set", raw)
## Running the MATLAB cleanline version
script = """
EEG = pop_loadset( 'test_raw.set', '/home/christian/Desktop/test_data/');
%EEG2 = cleanline(EEG);
EEG2 = cleanline(EEG, 'Bandwidth',2, ...
'SignalType','Channels','ComputeSpectralPower',true, ...
'LineFrequencies',[60 120] ,'NormalizeSpectrum',false, ...
'LineAlpha',0.01,'PaddingFactor',2,'PlotFigures',false, ...
'ScanForLines',true,'SmoothingFactor',100,'VerboseOutput',1, ...
'SlidingWinLength',EEG.pnts/EEG.srate,'SlidingWinStep',EEG.pnts/EEG.srate);
pop_saveset(EEG2, 'filename', 'after.set', 'filepath', '/home/christian/Desktop/test_data/', ...
'savemode', 'onefile');
"""
with open("cleanline_test.m","w+") as f:
f.write(script)
eng = matlab.engine.start_matlab()
eng.cleanline_test(nargout=0)
raw_after = mne.io.read_raw_eeglab("/home/christian/Desktop/test_data/after.set")
raw_after.set_montage(montage)
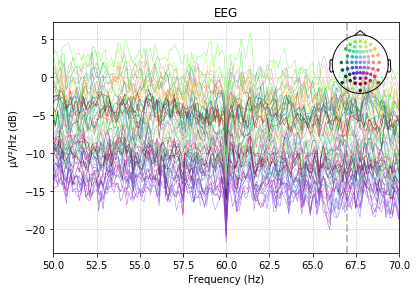
raw.plot_psd(fmin=50, fmax=70);raw_after.plot_psd(fmin=50, fmax=70);
## Running the pyprep version
eeg_index = mne.pick_types(raw.info, eeg=True, eog=False, meg=False)
ch_names = raw.info["ch_names"]
ch_names_eeg = list(np.asarray(ch_names)[eeg_index])
sample_rate = raw.info["sfreq"]
prep_params = {
"ref_chs": ch_names_eeg,
"reref_chs": ch_names_eeg,
"line_freqs": np.arange(60, sample_rate / 2, 60),
}
prep = PrepPipeline(raw.copy(), prep_params, montage)
prep.fit()
prep.raw.plot_psd(fmin=50, fmax=70);
It seems actually as if cleanline is doing nothing at all... but it is not throwing any error message. In any cases, it would be an understatement to say that I am not a cleanline expert. :)
from pyprep.
Just to chime in on this, the current notch filter implementation is consistently hard-crashing my Python session due to eating too much RAM. For context, this is me running a single recording (32 channel, 1000 Hz, ~40 minutes) through PyPREP.
After it crashed the first time I looked inside the PrepPipeline fit() function and started running it manually piece-by-piece. When I got to the notch_filter function, Python's RAM consumption was at ~2.9 GB according to Activity Monitor. After it had been running for 10-15 minutes, that number ballooned to 16 GB, then to 30 GB, eventually reaching ~55 GB (!!!) before macOS presumably killed it:
>>> EEG_raw = raw_copy.get_data() * 1e6
>>> EEG_new = removeTrend(EEG_raw, sample_rate=raw.info["sfreq"])
>>> EEG_clean = mne.filter.notch_filter(
... EEG_new,
... Fs=raw.info["sfreq"],
... freqs=linenoise,
... method="spectrum_fit",
... mt_bandwidth=2,
... p_value=0.01,
... )
Killed: 9
(eeg_env) bash-3.2$
I'll give the windowed mode PR a shot and see if that fixes it.
from pyprep.
how do you measure the RAM consumption? Do you have a code snippet so I can reproduce this and play around a bit?
I don't (although Im sure there are some python modules for this), I just have a log where I see the errors obtained , for example when trying to run with the notch filter I would get this sometimes:
2020-03-30 12:21:50,546 INFO root Traceback (most recent call last):
File "y:/code/preprocessing-flow/full_flow_db.py", line 307, in <module>
prep.fit()
File "Y:\code\preprocessing-flow\pyprep\prep_pipeline.py", line 72, in fit
p_value=0.01,
File "<C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\mne\externals\decorator.py:decorator-gen-103>", line 2, in notch_filter
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\mne\utils\_logging.py", line 90, in wrapper
return function(*args, **kwargs)
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\mne\filter.py", line 1190, in notch_filter
p_value, picks, n_jobs, copy)
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\mne\filter.py", line 1228, in _mt_spectrum_proc
threshold)
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\mne\filter.py", line 1323, in _mt_spectrum_remove
datafit = np.sum(np.atleast_2d(fits), axis=0)
File "<__array_function__ internals>", line 6, in atleast_2d
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\numpy\core\shape_base.py", line 123, in atleast_2d
ary = asanyarray(ary)
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\numpy\core\_asarray.py", line 138, in asanyarray
return array(a, dtype, copy=False, order=order, subok=True)
MemoryError: Unable to allocate 9.17 GiB for an array with shape (3648, 337360) and data type float64
Another one (similar head but not exactly the same though):
File "C:\Users\user\Anaconda3\envs\eeg\lib\site-packages\scipy\fft\_pocketfft\basic.py", line 58, in r2c
return pfft.r2c(tmp, (axis,), forward, norm, None, workers)
MemoryError: Unable to allocate 9.34 GiB for an array with shape (1119, 559873) and data type complex128
a single dataset? or all your data? Can you specify? This sounds awfully long 😬
Yes, on a single eeg record with about 10 minutes of activity recorded at 1000Hz with 58 channels. It takes awfully long to process each.
- Without the multitaper filter each record takes about 1 hour on the whole PREP.
- With the multitaper they will take about 1 hour and a half or more but rarely 2 hours.
So you gotta imagine how it is to process 70 eeg files...
In matlab it takes quite long too, I would guess about the same but I have never had memory errors.
from pyprep.
Yes in my limited experience it takes forever, and usually does not actually remove the noise. It could be (probably is) a bug in our implementation rather than a problem with the method, but I'm not sure if anyone will know how to fix it
from pyprep.
it seems the agreement between the matlab prep multitaper filter and the notch spectrum fit of mne is really high , like almost identical
that's additionally surprising because in MNE, it seems like the "pure" chronux Matlab toolbox functionality was copied (see docstr in mne filter.py)
Yet in the Matlab PREP, they are using the EEGLAB "cleanline" plugin (read "line noise removal" section in the prep paper), which is just built upon chronux (...but extends it): https://www.nitrc.org/projects/cleanline/
so it's surprising that the MNE port of some old chronux routine corresponds to the PREP wrapper around Cleanline, which extends Chronux.
from pyprep.
The way cleanline and mne work in this respect should differ - cleanline does not fit spectrum on the whole raw file, but does it in a moving-window fashion, then interpolates between windows (with linear or nonlinear interpolation - I don't remember what is the default). Cleanline also tests for significant presence of line noise frequency before trying to remove it. At least it worked this way some years ago when I was using it from EEGLAB.
from pyprep.
Yes the moving window approach was never implemented in MNE. I've been meaning to add a generic overlap-add processing mechanism for MNE for a while (I need it anyway), if I did that it might make it pretty easy to process in windows.
If that's "only" what's required to get it working, then it indeed might be salvagable and more usable at the MNE end!
from pyprep.
Feel free to try mne-tools/mne-python#7609, it implements windowed mode. Help appreciated fixing it as it still does not work very well for MEG data with line noise that I tested briefly at least...
from pyprep.
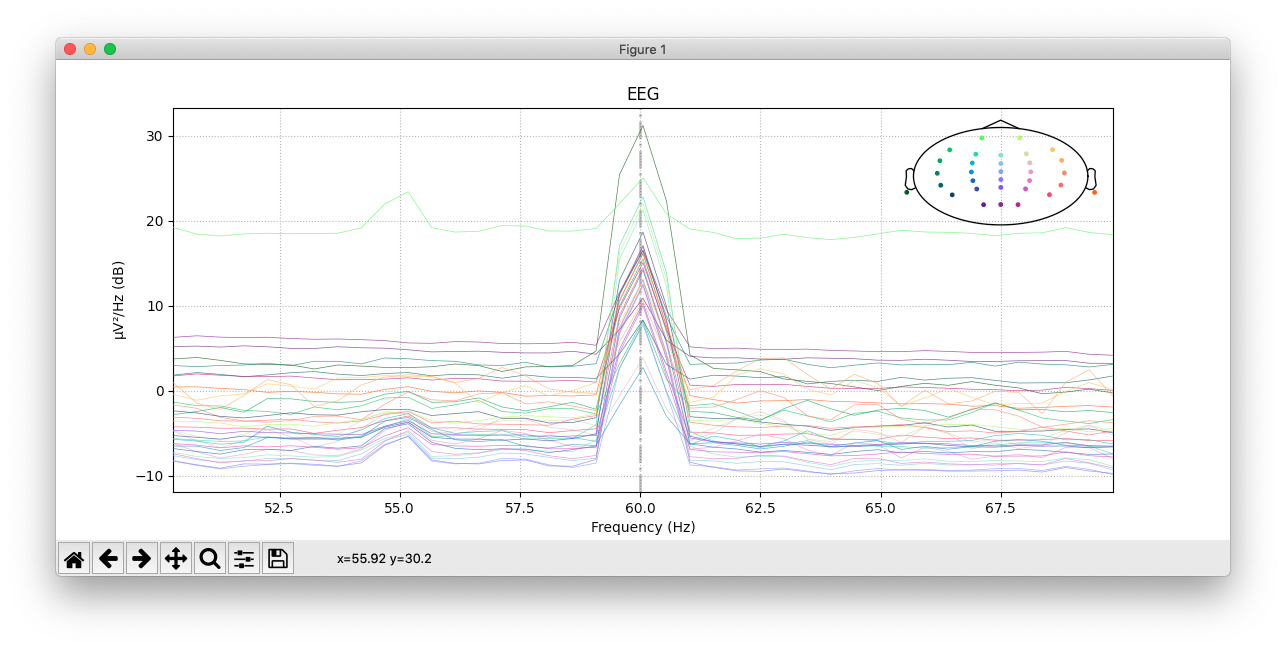
@yjmantilla Just did, it finishes fine now! Only takes a few minutes, with minimal RAM consumption. It doesn't look like it's 100% doing the right thing for my file though, but I'm not sure if that's on the implementation or on my settings.
This is the PSD pre-filter:
And this is the PSD after the windowed notch_filter:
I pretty much just used the code from fit() and added the filter_length="10s to the notch_filter arguments:
linenoise = np.arange(60, sample_rate / 2, 60)
EEG_raw = raw_copy.get_data() * 1e6
EEG_new = removeTrend(EEG_raw, sample_rate=raw.info["sfreq"])
EEG_clean = mne.filter.notch_filter(
EEG_new,
Fs=raw.info["sfreq"],
freqs=linenoise,
filter_length="10s",
method="spectrum_fit",
mt_bandwidth=2,
p_value=0.01,
)
EEG_final = EEG_raw - EEG_new + EEG_clean
raw_copy._data = EEG_final * 1e-6from pyprep.
@a-hurst My pleasure! I resisted for long to install MATLAB and I finally decided that it was time to compromise on my principles ;)
@sappelhoff Well, you already have the figures for cleanline and pyprep. Here is for MNE alone:
# Hugly hack because it was crashing with the raw directly. Something about picking
# indexes out of bound.
data_notched = mne.filter.notch_filter(
raw.get_data(),
Fs=raw.info["sfreq"],
freqs=[60.0, 120.0],
method="spectrum_fit",
mt_bandwidth=2,
p_value=0.01,
filter_length='10s'
)
raw_notched = mne.io.RawArray(data_notched, raw.info)
raw_notched.plot_psd(fmin=50, fmax=70);
Now, it really looks as if cleanline is doing nothing at all. I don't know why, when I look at the code, it seems fine and it does not throw any error. The diff looks like just numerical rounding errors...
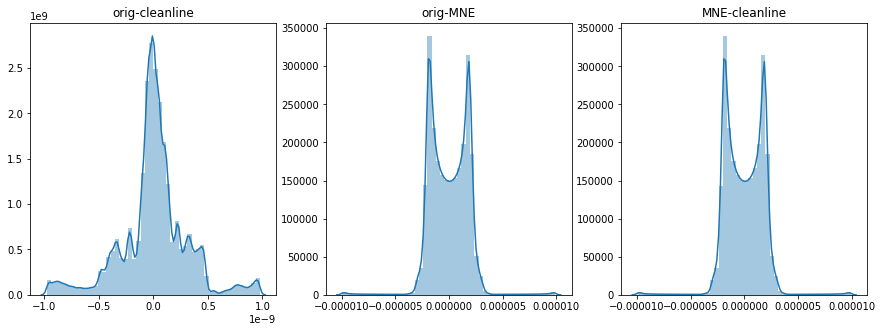
import seaborn as sns
import matplotlib.pyplot as plt
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
sns.distplot(raw.get_data() - raw_after.get_data(), ax=axes[0])
axes[0].set_title("orig-cleanline")
sns.distplot(raw.get_data() - raw_notched.get_data(), ax=axes[1])
axes[1].set_title("orig-MNE")
sns.distplot(raw_after.get_data() - raw_notched.get_data(), ax=axes[2])
axes[2].set_title("MNE-cleanline")
from pyprep.
Indeed it is doing nothing:
>> EEG = pop_loadset( 'test_raw.set', '/home/christian/Desktop/test_data/');
pop_loadset(): loading file /home/christian/Desktop/test_data/test_raw.set ...
>> EEG2 = cleanline(EEG, 'Bandwidth',2, ...
'SignalType','Channels','ComputeSpectralPower',true, ...
'LineFrequencies',[60 120] ,'NormalizeSpectrum',false, ...
'LineAlpha',0.01,'PaddingFactor',2,'PlotFigures',false, ...
'ScanForLines',true,'SmoothingFactor',100,'VerboseOutput',1, ...
'SlidingWinLength',EEG.pnts/EEG.srate,'SlidingWinStep',EEG.pnts/EEG.srate);
Welcome to the CleanLine line noise removal toolbox!
CleanLine is written by Tim Mullen ([email protected]) and uses multi-taper routines modified from the Chronux toolbox (www.chronux.org)
Tsk Tsk, you've allowed your data to get very dirty!
Let's roll up our sleeves and do some cleaning!
Today we're going to be cleaning your Channels
[!] Please note that because the selected window length does not divide the data length,
0 seconds of data at the end of the record will not be cleaned.
Multi-taper parameters follow:
Time-bandwidth product: 120
Number of tapers: 239
Number of FFT points: 131072
I'm going try to remove lines at these frequencies: [60 120] Hz
I'm going to scan the range +/-1 Hz around each of the above frequencies for the exact line frequency.
I'll do this by selecting the frequency that maximizes Thompson's F-statistic above a threshold of p=0.01.
OK, now stand back and let The Maid show you how it's done!
Converting spectra to dB...
>> max(max(EEG.data - EEG2.data))
ans =
single
0
I must say that I feel no compulsion to debug cleanline at the time...
from pyprep.
I don't get it --> pyprep currently consumes a lot of run during a single MNE mne.filter.notch_filter filter call? How can that be?
from pyprep.
Yes exactly, in a single filter call it consumes a lot of ram.
At least speaking for the cleanline method (the filter in matlab) it has to estimate the phase and amplitude of a deterministic sinusoid for each time window for each channel for each expected line frequency harmonics. It does this in many iterations until the power of that sinusoid is indistinguishable from noise. I noticed this takes quite a lot of time ie 30 min to 1 hour on my eeg records (not sure of the ram consumption in matlab though).
I dont know if the mne multitaper notch filter works exactly the same as the cleanline method of prep but given that in matlab the filter takes that much time it does not surprise me that in mne it requires a lot of ram (and time too).
from pyprep.
Yes exactly, in a single filter call it consumes a lot of ram.
how do you measure the RAM consumption? Do you have a code snippet so I can reproduce this and play around a bit?
I noticed this takes quite a lot of time ie 30 min to 1 hour on my eeg records
a single dataset? or all your data? Can you specify? This sounds awfully long 😬
from pyprep.
So you gotta imagine how it is to process 70 eeg files...
that sounds just unacceptable ...
I don't have time to go down this rabbit hole right now unfortunately, but we should keep it on the list and do more digging and testing of this.
from pyprep.
Yes in my limited experience it takes forever, and usually does not actually remove the noise. It could be (probably is) a bug in our implementation rather than a problem with the method, but I'm not sure if anyone will know how to fix it
That does makes me wonder. From the tests I made in #21 and the ones done by the neuro data design team of Stanford @AamnaLawrence @Nick3151 it seems the agreement between the matlab prep multitaper filter and the notch spectrum fit of mne is really high , like almost identical. So it would be weird for there to be a bug (or it is in both implementations?). Regarding the effectiveness of it I haven't really made any observations. In any case this is mostly relevant for ERP folks, most others will probably be happy with normal notch.
from pyprep.
The way cleanline and mne work in this respect should differ - cleanline does not fit spectrum on the whole raw file, but does it in a moving-window fashion, then interpolates between windows (with linear or nonlinear interpolation - I don't remember what is the default). Cleanline also tests for significant presence of line noise frequency before trying to remove it. At least it worked this way some years ago when I was using it from EEGLAB.
@sappelhoff @larsoner @mmagnuski It is possible that the signals we tested the spectrum fit against clean line didn't have that much line noise so in the end they both didn't affect the signal thus ending in an extremely similar result. So a good sets of known tests would be helpful and porting it to a window-based would be the solution to the ram problem.
from pyprep.
Agreed we need good test files. It sounds like the default window length for cleanline is 4 sec (at least according to this wiki
That wiki also talks about some failure modes of cleanline due to non-stationarity. Of course the failure mode there has to do with AM on the 60 Hz that is above 0.25 Hz (what is presumably the limit of cleanline operating on 4 sec windows). To remove that with traditional filtering you'd need to notch with a width at least twice as wide as the AM freq -- eyeballing it, it looks like there's something like a ~4 Hz AM on that so you need a pretty wide notch (10 Hz probably?) to get rid of it. So maybe it's not a critical failure mode for most datasets, which don't suffer from such bad line freq AM...
from pyprep.
That wiki also talks about some failure modes of cleanline due to non-stationarity.
Thats really interesting, so it could be that both prep's clean line and mne spectrum fit were unsuccessful to eliminate anything and thus arrive at similar results.
I did some quick tests based on simulated sources (SourceSimulator class) and the projection of it to EEG. I have found the following:
-
Its easy for the filter to miss the frequencies if they are not exactly what you put in the freqs argument. I tested this with real freq at 61 Hz and 60 in the argument. Even by playing with the notch widths arguments I couldn't really get noticeably better.
-
If one sets freqs to None it does attenuate the line freqs but may attenuate some oscillators present on the data (as in my case I put a bunch of oscillators in the sources). The attenuation is not on the level of what a normal notch filter would achieve.
from pyprep.
@a-hurst please do and report back to us. I have been busy lately but will be soon a little more free to test that windowed mode.
A new comment in mne-tools/mne-python#7609 suggest it could work ok
from pyprep.
Yeah it seems like the behaviour is still like before, sometimes it works and others it doesnt. I will see what I can do in about a week.
from pyprep.
Thanks @a-hurst :-) also tagging @christian-oreilly here because he may or may not be aware of this thread.
Looks like it's working --- but not working well. I.e., the peak is reduced but still there 😕
Anyhow, PRs to pyprep are very welcome
from pyprep.
Can someone test this file with another implementation of this filtering technique? It's entirely possible there is some bug in MNE. If someone can, say, crop the file to 10 seconds and process with MNE and then the other software (Chronux?) and see that MNE doesn't remove the line noise but the other software does, it might not be too much work from there to dig into the two sets of code to figure out where MNE's bug is.
from pyprep.
@larsoner I'd be happy to (I could also just share the file, since it'll hopefully make its way to OSF eventually), but unfortunately my MATLAB experience is limited and I don't have a licence.
Do you (or anyone else) know off-hand of any example EEGLAB PREP scripts I could adapt to use its CleanLine implementation on my file? From what I've read, EEGLAB works fine in GNU Octave but the GUI is broken, so I'd have to use a script instead of a point-and-click guide (better reproducability anyway).
from pyprep.
@christian-oreilly looks great, thanks for posting this! Saves me the headache of trying to spend a day learning Octave and EEGLAB.
This sort of confirms my suspicion that it's my particular file being a problem: if you look at my post-filter PSD you can see that several channels are completely flat at 60 Hz despite the majority still having a peak, which maps on to the pre-filter PSD where you can see the magnitude of the line noise is stronger for some channels than others. My guess is that the issue probably has to do with very bad channels in the data not being dropped before filtering, though I don't know enough about the algorithms to know whether that makes sense.
Regardless, it looks like the windowed "spectrum_fit" does work properly, and it at least runs instead of crashing after taking up 5x my system RAM, so I'd say it's definitely an improvement and should be merged.
from pyprep.
@christian-oreilly could you also show us the MNE result without running full pyprep? So that we can compare:
- cleanline
- MNE
- pyprep
? :-)
from pyprep.
gosh that cleanline is talkative :-)
Thanks a lot @christian-oreilly
super weird that it doesn't seem to work 😕 I'd have thought this is being used regularly and that a bug would become obvious fast.
from pyprep.
'SlidingWinLength',EEG.pnts/EEG.srate,'SlidingWinStep',EEG.pnts/EEG.srate);
This doesn't look like it's going to use 10 second windows, but rather (more or less) srate windows. The window length is already in seconds according to the cleanline docs so these should probably both just be 10.
This is also suspicious:
Number of tapers: 239
from pyprep.
There has been an update a few days ago apparently ... maybe worth looking into (or not): sccn/cleanline@294a617
from pyprep.
I would propose that we should get things equivalent with the "old version" first, then move to this new one.
Also that license on that repo is GPL so we can't use that particular version anyway. I think EEGLAB has BSD nowadays (?) in which case we might be able to look there.
from pyprep.
@larsoner Sorry, I contributed to the confusion. I initially used a 10s file but then I changed it for 120s (because of some issues I had). Not sure how we get "Time-bandwidth product: 120" (In my simplistic mind "time-bandwith product" would mean "the product of the the bandwidth (=2) by the time duration (=120)" which would equals 240, but I guess life is more complicated than that). Also not sure what is the formula that relate these different parameters to the number of tapers... but in any case, the duration is 120s here.
from pyprep.
Related Issues (20)
- Use the MNE logger to set the verbosity HOT 6
- Make pyprep load MNE raw data for object, if not already loaded HOT 3
- filt vs filtfilt (MATLAB implementation)
- Add a "reject_by_annotation" parameter for finding bad channels HOT 1
- pyprep.utils._filter_design throws error for large sampling rates HOT 4
- Passing a custom montage to the PrepPipeline(raw, montage=?)? HOT 5
- Saving prep.raw give error " ValueError: Measurement infos are inconsistent for dig" HOT 4
- List of channel names causes TypeError in find_bad_by_ransac HOT 2
- migrate from `.zenodo.json` to `CITATION.cff` HOT 1
- update issue/pr templates HOT 1
- Computation of window size based on cutoff frequency in local detrend method HOT 1
- Question: using pyprep.NoisyChannels on numpy arrays HOT 1
- Issue with RawBrainVision HOT 2
- find_all_bads throws inf/nan error after filtering/detrending? HOT 12
- Documentation for Noisy Channels Algorithms' stand-alone use HOT 1
- [Feature suggestion] Allow for relevant annotation selection during processing.
- How to include the prep output in my preprocessing pipeline? HOT 1
- New release? HOT 4
- Possible to add an argument in find_bad_by_nan_flat() to change FLAT_THRESHOLD ? HOT 2
- Add to NoisyChannel a bad_by_psd method to tackle low-frequency artefacts HOT 8
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.






from pyprep.