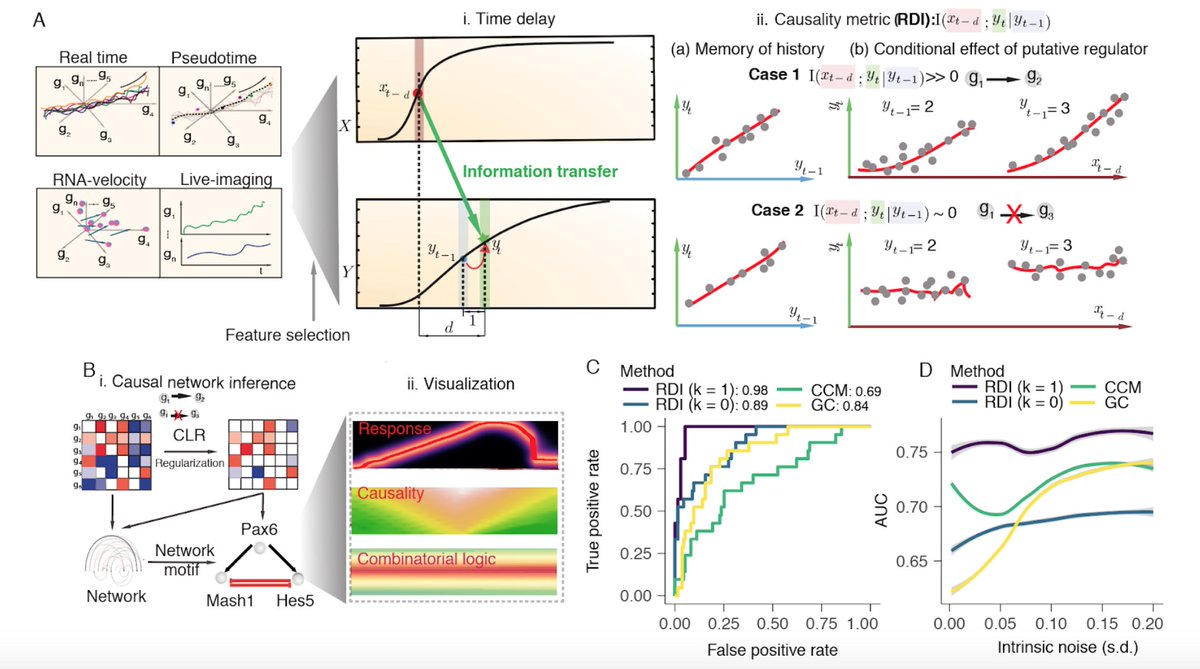
Single-cell transcriptome sequencing now routinely samples thousands of cells, potentially providing enough data to reconstruct causal gene regulatory networks from observational data. Here, we developed Scribe, a toolkit for detecting and visualizing causal regulations, and explore the potential for single-cell experiments to power network reconstruction. Scribe employs Restricted Directed Information to determine causality by estimating the strength of information transferred from a potential regulator to its downstream target by taking advantage of time-delays. We apply Scribe and other leading approaches for network reconstruction to several types of single-cell measurements and show that there is a dramatic drop in performance for "pseudotime” ordered single-cell data compared to live imaging data. We demonstrate that performing causal inference requires temporal coupling between measurements. We show that methods such as “RNA velocity” restore some degree of coupling through an analysis of chromaffin cell fate commitment. These analyses therefore highlight an important shortcoming in experimental and computational methods for analyzing gene regulation at single-cell resolution and point the way towards overcoming it.
This was a serious bug in causal_net_dynamics_coupling function which
leads to incorrect calculation of causality score, if you used it
previously please update your analysis wit the fix (May 13, 2020).
Note that this is our first alpha version of Scribe (as of Aug. 11th, 2019) python package. Scribe is still under active development. Stable version of Scribe will be released when it is ready. Until then, please use Scribe with caution. We welcome any bugs reports (via GitHub issue reporter) and especially code contribution (via GitHub pull requests) of Scribe from users to make it an accessible, useful and extendable tool. For discussion about different usage cases, comments or suggestions related to our manuscript and questions regarding the underlying mathematical formulation of Scribe, we provided a google group goolge group. Scribe developers can be reached by [email protected]. To install the newest version of Scribe, you can git clone our repo and then use::
pip install directory_to_Scribe_py_repo/Alternatively, You can install Scribe from source, using the following script:
pip install git+https://github.com:aristoteleo/Scribe-pyXiaojie Qiu, Arman Rahimzamani, Li Wang, Qi Mao, Timothy Durham, Jose L McFaline-Figueroa, Lauren Saunders, Cole Trapnell, Sreeram Kannan (2018): Towards inferring causal gene regulatory networks from single cell expression measurements. BioRxiv
Cell Systems link: https://www.sciencedirect.com/science/article/abs/pii/S2405471220300363 (downloadable from here: http://cole-trapnell-lab.github.io/papers/qiu-scribe/)
biorxiv link: https://www.biorxiv.org/content/early/2018/09/25/426981
twitter link: https://twitter.com/coletrapnell/status/1044986820520435712
A R version of this package is available at: https://github.com/cole-trapnell-lab/Scribe. Note that I have graduated Cole's lab and won't maintain this package anymore. If anyone wants to maintain it and keep it updated. Please let me know (email: [email protected]).
I am recently working on developing a new framework that tries to go beyond RNA velocity to map the full vector field of single cells. You may find this project interesting (https://github.com/aristoteleo/dynamo-release). In a month or two, Scribe will be fully integrated with Dynamo, so stayed tuned.
If you want to contribute to the development of Scribe, please check out CONTRIBUTION instruction: Contribution
The documentation of Scribe package is available at readthedocs