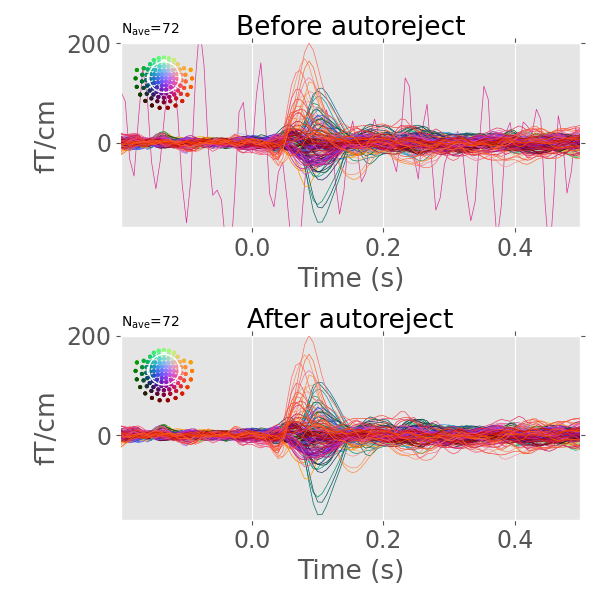
This is a library to automatically reject bad trials and repair bad sensors in magneto-/electroencephalography (M/EEG) data.
The documentation can be found under the following links:
- for the stable release
- for the latest (development) version
We recommend the Anaconda Python distribution
and a Python version >= 3.9.
We furthermore recommend that you install autoreject into an isolated
Python environment.
To obtain the stable release of autoreject, you can use pip:
pip install -U autoreject
Or conda:
conda install -c conda-forge autoreject
If you want the latest (development) version of autoreject, use:
pip install https://github.com/autoreject/autoreject/archive/refs/heads/main.zip
To check if everything worked fine, you can do:
python -c 'import autoreject'
and it should not give any error messages.
Below, we list the dependencies for autoreject.
All required dependencies are installed automatically when you install autoreject.
mne(>=1.5.0)numpy(>=1.21.2)scipy(>=1.7.1)scikit-learn(>=1.0.0)joblibmatplotlib(>=3.5.0)
Optional dependencies are:
openneuro-py(>= 2021.10.1, for fetching data from OpenNeuro.org)
The easiest way to get started is to copy the following three lines of code in your script:
>>> from autoreject import AutoReject
>>> ar = AutoReject()
>>> epochs_clean = ar.fit_transform(epochs) # doctest: +SKIPThis will automatically clean an epochs object read in using MNE-Python. To get the rejection dictionary, simply do:
>>> from autoreject import get_rejection_threshold
>>> reject = get_rejection_threshold(epochs) # doctest: +SKIPWe also implement RANSAC from the PREP pipeline (see PyPREP for a full implementation of the PREP pipeline). The API is the same:
>>> from autoreject import Ransac
>>> rsc = Ransac()
>>> epochs_clean = rsc.fit_transform(epochs) # doctest: +SKIPFor more details check out the example to automatically detect and repair bad epochs.
Please use the GitHub issue tracker to report bugs.
[1] Mainak Jas, Denis Engemann, Federico Raimondo, Yousra Bekhti, and Alexandre Gramfort, "Automated rejection and repair of bad trials in MEG/EEG." In 6th International Workshop on Pattern Recognition in Neuroimaging (PRNI), 2016.
[2] Mainak Jas, Denis Engemann, Yousra Bekhti, Federico Raimondo, and Alexandre Gramfort. 2017. "Autoreject: Automated artifact rejection for MEG and EEG data". NeuroImage, 159, 417-429.