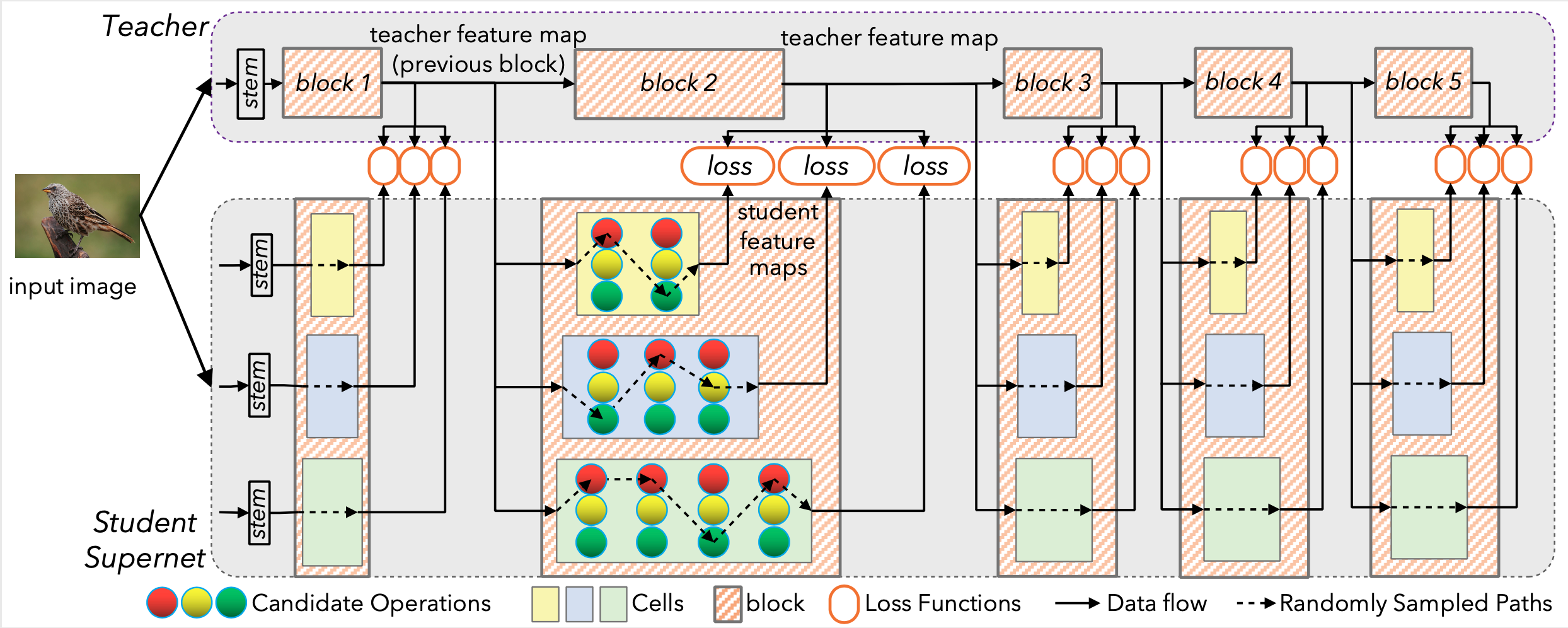
Have you encountered these issues before or have any idea how to fix them? TIA.
12/28 07:27:02 AM WORLD_SIZE in os.environ is 1
12/28 07:27:02 AM Namespace(amp=False, batch_size=64, color_jitter=0.4, cooldown_epochs=0, data_config=None, datadir='/home/ubuntu/workspace/datasets/ILSVRC2012/', dataset='imagenet', decay_epochs=1, decay_rate=0.9, distill_last_stage=True, distributed=False, eval_intervals=2, eval_metric='prec1', eval_mode=False, exp_dir='', feature_train=True, guide_input=True, guide_loss_fn='mse', hyperparam_config=None, img_size=224, index='', init_classifier=False, interpolation='', label_train=False, local_rank=0, log_interval=50, loss_weight=[0.5, 0.5], lr=[0.002, 0.005, 0.005, 0.005, 0.005, 0.002], mean=None, min_lr=1e-08, mixup=0.0, mixup_off_epoch=0, model_ema=False, model_ema_decay=0.9998, model_ema_force_cpu=False, model_pool='', momentum=0.9, num_classes=1000, num_gpu=1, opt='adam', opt_eps=1e-08, output='', potential_eval_times=20, prefetcher=True, pretrain=False, print_detail=True, recovery_interval=0, remode='pixel', reprob=0.5, reset_after_stage=False, reset_bn_eval=True, resume='', reverse_train=False, save_images=False, save_last_feature=True, sched='step', seed=42, separate_train=False, smoothing=0.1, stage_num=6, start_epoch=None, start_stage=None, std=None, step_epochs=20, sync_bn=False, test_dispatch='', top_model_num=3, train_mode=False, update_frequency=1, warmup_epochs=0, warmup_lr=0.001, weight_decay=0.0001, workers=4)
12/28 07:27:02 AM Training with a single process on 1 GPUs.
12/28 07:27:04 AM Data processing configuration for current model + dataset:
12/28 07:27:04 AM input_size: (3, 224, 224)
12/28 07:27:04 AM interpolation: bicubic
12/28 07:27:04 AM mean: (0.485, 0.456, 0.406)
12/28 07:27:04 AM std: (0.229, 0.224, 0.225)
12/28 07:27:04 AM crop_pct: 0.875
12/28 07:27:06 AM NVIDIA APEX installed. AMP off.
12/28 07:27:32 AM
Train: stage 0, epoch 1, step [ 0/20018] Loss: 109.597771 (109.5978) Time: 2.011s, 31.82/s LR: 1.800e-03 Data & Guide Time: 1.644
GuideMean: -0.64644 GuideStd: 10.40032 OutMean: 0.00000 (0.00000) OutStd: 0.99985 (0.99985) Dist_Mean: 0.64644 (0.64644)
GRLoss: 1.00459 (1.00459) CLLoss: 0.79709 (0.79709) KLCosLoss: 0.57991 (0.57991)
FeatureLoss: 0.00000 (0.00000) Top1Acc: 0.00000(0.00000)
Relative MSE loss: 1.01323(1.01323)
.....
12/29 06:58:47 AM Random Test: stage 0, epoch 20 Loss: 20.4754 Prec@1: 0.0000 Time: 0.216s, 74.05/s
12/29 06:58:48 AM Current checkpoints:
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-6.pth.tar', 19.889211503295897)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-14.pth.tar', 19.960276111450195)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-4.pth.tar', 19.97588088684082)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-16.pth.tar', 20.030977337646483)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-8.pth.tar', 20.106792897033692)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-10.pth.tar', 20.107453624572752)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-12.pth.tar', 20.242049604492188)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-18.pth.tar', 20.277006747436523)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-2.pth.tar', 20.39269996520996)
('./output/test/adam-step-ep20-lr0.002-bs64-20201228-072702/checkpoint-0-20.pth.tar', 20.47537907836914)
Traceback (most recent call last):
File "train.py", line 273, in <module>
main()
File "train.py", line 268, in main
writer=writer)
File "/home/ubuntu/workspace/repos/DNA/searching/dna/distill_train.py", line 100, in distill_train
reset_data=reset_data)
File "/home/ubuntu/workspace/repos/DNA/searching/dna/distill_train.py", line 695, in _potential
for layer in supernet.module.modules():
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages/torch/nn/modules/module.py", line 576, in __getattr__
type(self).__name__, name))
AttributeError: 'StudentSuperNet' object has no attribute 'module'
Traceback (most recent call last):
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/runpy.py", line 193, in _run_module_as_main
"__main__", mod_spec)
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/runpy.py", line 85, in _run_code
exec(code, run_globals)
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages/torch/distributed/launch.py", line 263, in <module>
main()
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages/torch/distributed/launch.py", line 259, in main
cmd=cmd)
subprocess.CalledProcessError: Command '['/home/ubuntu/anaconda3/envs/pytorch_p36/bin/python', '-u', 'train.py', '--local_rank=0']' returned non-zero exit status 1.
12/30 05:01:12 AM WORLD_SIZE in os.environ is 4
12/30 05:01:12 AM Namespace(amp=False, batch_size=64, color_jitter=0.4, cooldown_epochs=0, data_config=None, datadir='/home/ubuntu/workspace/datasets/ILSVRC2012/', dataset='imagenet', decay_epochs=1, decay_rate=0.9, distill_last_stage=True, distributed=False, eval_intervals=2, eval_metric='prec1', eval_mode=False, exp_dir='', feature_train=True, guide_input=True, guide_loss_fn='mse', hyperparam_config=None, img_size=224, index='', init_classifier=False, interpolation='', label_train=False, local_rank=0, log_interval=50, loss_weight=[0.5, 0.5], lr=[0.002, 0.005, 0.005, 0.005, 0.005, 0.002], mean=None, min_lr=1e-08, mixup=0.0, mixup_off_epoch=0, model_ema=False, model_ema_decay=0.9998, model_ema_force_cpu=False, model_pool='', momentum=0.9, num_classes=1000, num_gpu=1, opt='adam', opt_eps=1e-08, output='', potential_eval_times=20, prefetcher=True, pretrain=False, print_detail=True, recovery_interval=0, remode='pixel', reprob=0.5, reset_after_stage=False, reset_bn_eval=True, resume='', reverse_train=False, save_images=False, save_last_feature=True, sched='step', seed=42, separate_train=False, smoothing=0.1, stage_num=6, start_epoch=None, start_stage=None, std=None, step_epochs=20, sync_bn=False, test_dispatch='', top_model_num=3, train_mode=False, update_frequency=1, warmup_epochs=0, warmup_lr=0.001, weight_decay=0.0001, workers=4)
12/30 05:01:12 AM Training in distributed mode with multiple processes, 1 GPU per process. CUDA 2, Process 2, total 4.
12/30 05:01:12 AM Training in distributed mode with multiple processes, 1 GPU per process. CUDA 3, Process 3, total 4.
12/30 05:01:13 AM Training in distributed mode with multiple processes, 1 GPU per process. CUDA 1, Process 1, total 4.
12/30 05:01:13 AM Training in distributed mode with multiple processes, 1 GPU per process. CUDA 0, Process 0, total 4.
12/30 05:01:15 AM Data processing configuration for current model + dataset:
12/30 05:01:15 AM input_size: (3, 224, 224)
12/30 05:01:15 AM interpolation: bicubic
12/30 05:01:15 AM mean: (0.485, 0.456, 0.406)
12/30 05:01:15 AM std: (0.229, 0.224, 0.225)
12/30 05:01:15 AM crop_pct: 0.875
12/30 05:01:18 AM NVIDIA APEX installed. AMP off.
ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@ERROR: Unexpected segmentation fault encountered in worker.
^@Traceback (most recent call last):
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/runpy.py", line 193, in _run_module_as_main
"__main__", mod_spec)
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/runpy.py", line 85, in _run_code
exec(code, run_globals)
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages/torch/distributed/launch.py", line 263, in <module>
main()
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages/torch/distributed/launch.py", line 256, in main
process.wait()
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/subprocess.py", line 1477, in wait
(pid, sts) = self._try_wait(0)
File "/home/ubuntu/anaconda3/envs/pytorch_p36/lib/python3.6/subprocess.py", line 1424, in _try_wait
(pid, sts) = os.waitpid(self.pid, wait_flags)