This pile of code was used to simulate biological creatures that evolve through natural selection. The results of the experiments are summarized in this YouTube video:
"I programmed some creatures. They evolved."
https://www.youtube.com/watch?v=N3tRFayqVtk
This code lacks a friendly interface, so compiling and executing the program may require attention to details. If you ask questions in the Issues, I'll try to help if I can.
- Code walkthrough
- Main data structures
- Config file
- Program output
- Main program loop
- Sensory inputs and action outputs
- Basic value types
- Pheromones
- Useful utility functions
- Installing the code
- Building the executable
- System requirements
- Compiling
- Bugs
- Execution
- Tools directory
- Build log
The code in the src directory compiles to a single console program named biosim4. When it is invoked, it will read parameters from a config file named biosim4.ini by default. A different config file can be specified on the command line.
The simulator will then configure a 2D arena where the creatures live. Class Grid (see grid.h and grid.cpp) contains a 2D array of 16-bit indexes, where each nonzero index refers to a specific individual in class Peeps (see below). Zero values in Grid indicate empty locations. Class Grid does not know anything else about the world; it only stores indexes to represent who lives where.
The population of creatures is stored in class Peeps (see peeps.h and peeps.cpp). Class Peeps contains all the individuals in the simulation, stored as instances of struct Indiv in a std::vector container. The indexes in class Grid are indexes into the vector of individuals in class Peeps. Class Peeps keeps a container of struct Indiv, but otherwise does not know anything about the internal workings of individuals.
Each individual is represented by an instance of struct Indiv (see indiv.h and indiv.cpp). Struct Indiv contains an individual's genome, its corresponding neural net brain, and a redundant copy of the individual's X,Y location in the 2D grid. It also contains a few other parameters for the individual, such as its "responsiveness" level, oscillator period, age, and other personal parameters. Struct Indiv knows how to convert an individual's genome into its neural net brain at the beginning of the simulation. It also knows how to print the genome and neural net brain in text format to stdout during a simulation. It also has a function Indiv::getSensor() that is called to compute the individual's input neurons for each simulator step.
All the simulator code lives in the BS namespace (short for "biosim".)
The config file, named biosim4.ini by default, contains all the tunable parameters for a simulation run. The biosim4 executable reads the config file at startup, then monitors it for changes during the simulation. Although it's not foolproof, many parameters can be modified during the simulation run. Class ParamManager (see params.h and params.cpp) manages the configuration parameters and makes them available to the simulator through a read-only pointer provided by ParamManager::getParamRef().
See the provided biosim4.ini for documentation for each parameter. Most of the parameters in the config file correspond to members in struct Params (see params.h). A few additional parameters may be stored in struct Params. See the documentation in params.h for how to support new parameters.
Depending on the parameters in the config file, the following data can be produced:
-
The simulator will append one line to logs/epoch.txt after the completion of each generation. Each line records the generation number, number of individuals who survived the selection criterion, an estimate of the population's genetic diversity, average genome length, and number of deaths due to the "kill" gene. This file can be fed to tools/graphlog.gp to produce a graphic plot.
-
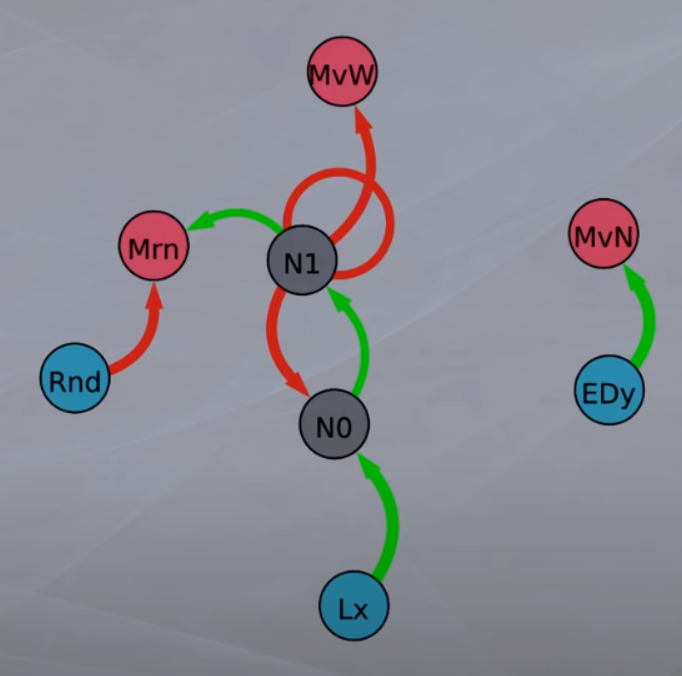
The simulator will display a small number of sample genomes at regular intervals to stdout. Parameters in the config file specify the number and interval. The genomes are displayed in hex format and also in a mnemonic format that can be fed to tools/graph-nnet.py to produce a graphic network diagram.
-
Movies of selected generations will be created in the images/ directory. Parameters in the config file specify the interval at which to make movies. Each movie records a single generation.
-
At intervals, a summary is printed to stdout showing the total number of neural connections throughout the population from each possible sensory input neuron and to each possible action output neuron.
The simulator starts with a call to simulator() in simulator.cpp. After initializing the world, the simulator executes three nested loops: the outer loop for each generation, an inner loop for each simulator step within the generation, and an innermost loop for each individual in the population. The innermost loop is thread-safe so that it can be parallelized by OpenMP.
At the end of each simulator step, a call is made to endOfSimStep() in single-thread mode (see endOfSimStep.cpp) to create a video frame representing the locations of all the individuals at the end of the simulator step. The video frame is pushed on to a stack to be converted to a movie later. Also some housekeeping may be done for certain selection scenarios. See the comments in endOfSimStep.cpp for more information.
At the end of each generation, a call is made to endOfGeneration() in single-thread mode (see endOfGeneration.cpp) to create a video from the saved video frames. Also a new graph might be generated showing the progress of the simulation. See endOfGeneraton.cpp for more information.
See the YouTube video (link above) for a description of the sensory inputs and action outputs. Each sensory input and each action output is a neuron in the individual's neural net brain.
The header file sensors-actions.h contains enum Sensor which enumerates all the possible sensory inputs and enum Action which enumerates all the possible action outputs. In enum Sensor, all the sensory inputs before the enumerant NUM_SENSES will be compiled into the executable, and all action outputs before NUM_ACTIONS will be compiled. By rearranging the enumerants in those enums, you can select a subset of all possible sensory and action neurons to be compiled into the simulator.
There are a few basic value types:
-
enum Compass represents eight-way directions with enumerants N=0, NE, E, SW, S, SW, W, NW, CENTER.
-
struct Dir is an abstract representation of the values of enum Compass.
-
struct Coord is a signed 16-bit integer X,Y coordinate pair. It is used to represent a location in the 2D world, or can represent the difference between two locations.
-
struct Polar holds a signed 32-bit integer magnitude and a direction of type Dir.
Various conversions and math are possible between these basic types. See unitTestBasicTypes.cpp for examples. Also see basicTypes.h for more information.
A simple system is used to simulate pheromones emitted by the individuals. Pheromones are called "signals" in simulator-speak (see signals.h and signals.cpp). Struct Signals holds a single layer that overlays the 2D world in class Grid. Each location can contain a level of pheromone (there's only a single kind of pheromone supported at present). The pheromone level at any grid location is stored as an unsigned 8-bit integer, where zero means no pheromone, and 255 is the maximum. Each time an individual emits a pheromone, it increases the pheromone values in a small neighborhood around the individual up to the maximum value of 255. Pheromone levels decay over time if they are not replenished by the individuals in the area.
The utility function visitNeighborhood() in grid.cpp can be used to execute a user-defined lambda or function over each location within a circular neighborhood defined by a center point and floating point radius. The function calls the user-defined function once for each location, passing it a Coord value. Only locations within the bounds of the grid are visited. The center location is included among the visited locations. For example, a radius of 1.0 includes only the center location plus four neighboring locations. A radius of 1.5 includes the center plus all the eight-way neighbors. The radius can be arbitrarily large but large radii require lots of CPU cycles.
Copy the directory structure to a location of your choice.
This code is known to run in the following environment:
- Ubuntu 21.04, 22.04, or Debian 10 (Buster)
- cimg-dev 2.4.5 or later
- libopencv-dev 3.2 or later
- gcc 8.3, 9.3 or 10.3
- python-igraph 0.8.3 (used only by tools/graph-nnet.py)
- gnuplot 5.2.8 (used only by tools/graphlog.gp)
The code also runs in distributions based on Ubuntu 20.04, but only if the default version of cimg-dev is replaced with version 2.8.4 or later.
You have several options:
The file named "biosim4.cbp" is a configuration file for the Code::Blocks IDE version 20.03.
A Makefile is provided which was created from biosim4.cbp with cbp2make. Possible make commands include:
- "make" with no arguments makes release and debug versions in ./bin/Release and ./bin/Debug
- "make release" makes a release version in ./bin/Release
- "make debug" makes a debug version in ./bin/Debug
- "make clean" removes the intermediate build files
A Dockerfile is provided which leverages the aforementioned Makefile.
To build a Docker environment in which you can compile the program:
docker build -t biosim4 .You can then compile the program with an ephemeral container:
docker run --rm -ti -v `pwd`:/app --name biosim biosim4 makeWhen you exit the container, the files compiled in your container files will persist in ./bin.
A CMakeList.txt file is provided to allow development, build, test, installation and packaging with the CMake tool chain and all IDE's that support CMake.
To build with cmake you need to install cmake. Once installed use the procedure below:
mkdir build
cd build
cmake ../
cmake --build ./To make a test installation and run the program:
mkdir build
cd build
cmake ../
cmake --build ./
mkdir test_install
cmake --install ./ --prefix ./test_install
cd test_install
./bin/biosim4To make a release package:
mkdir build
cd build
cmake ../
cmake --build ./
cpack ./If you try to compile the simulator under a distribution based on Ubuntu 20.04, you will encounter this bug in the version of CImg.h (package cimg-dev) provided by the package maintainer:
https://bugs.debian.org/cgi-bin/bugreport.cgi?bug=951965
In biosim4, CImg.h is used only as a convenient interface to OpenCV to generate movies of the simulated creatures in their 2D world. You have several choices if you want to proceed with Ubuntu 20.04:
-
You can strip out the code that generates the movies and just run the simulator without the movies. Most of that graphics code is in imageWriter.cpp and imageWriter.h.
-
You can upgrade your CImg.h to version 2.8.4 or later by installing the Ubuntu 22.04 cimg-dev package, For example:
cd /tmp && \
wget http://mirrors.kernel.org/ubuntu/pool/universe/c/cimg/cimg-dev_2.9.4+dfsg-3_all.deb -O cimg-dev_2.9.4+dfsg-3_all.deb && \
sudo apt install ./cimg-dev_2.9.4+dfsg-3_all.deb && \
rm cimg-dev_2.9.4+dfsg-3_all.deb;
- You could convert the CImg.h function calls to use OpenCV directly. Sorry I don't have a guide for how to do that.
Test everything is working by executing the Debug or Release executable in the bin directory with the default config file ("biosim4.ini"). e.g.:
./bin/Release/biosim4 biosim4.ini
You should have output something like:
Gen 1, 2290 survivors
If this works then edit the config file ("biosim4.ini") for the parameters you want for the simulation run and execute the Debug or Release executable. Optionally specify the name of the config file as the first command line argument, e.g.:
./bin/Release/biosim4 [biosim4.ini]
Note: If using docker,
docker run --rm -ti -v `pwd`:/app --name biosim biosim4 bashwill put you into
an environment where you can run the above and have all your files persist when you exit (using Ctrl-D).
tools/graphlog.gp takes the generated log file logs/epoch-log.txt and generates a graphic plot of the simulation run in images/log.png. You may need to adjust the directory paths in graphlog.gp for your environment. graphlog.gp can be invoked manually, or if the option "updateGraphLog" is set to true in the simulation config file, the simulator will try to invoke tools/graphlog.gp automatically during the simulation run. Also see the parameter named updateGraphLogStride in the config file.
tools/graph-nnet.py takes a text file (hardcoded name "net.txt") and generates a neural net connection diagram using igraph. The file net.txt contains an encoded form of one genome, and must be the same format as the files generated by displaySampleGenomes() in src/analysis.cpp which is called by simulator() in src/simulator.cpp. The genome output is printed to stdout automatically if the parameter named "displaySampleGenomes" is set to nonzero in the config file. An individual genome can be copied from that output stream and renamed "net.txt" in order to run graph-nnet.py.
Note: If using the docker run ... bash command, the presumed directory structure would necessitate the
following syntax:
gnuplot tools/graphlog.gp
cd tools && python3 graph-nnet.pyIn case it helps for debugging the build process, here is a build log from Code::Blocks running under Ubuntu 21.04:
-------------- Clean: Release in biosim4 (compiler: GNU GCC Compiler)---------------
Cleaned "biosim4 - Release"
-------------- Build: Release in biosim4 (compiler: GNU GCC Compiler)---------------
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/analysis.cpp -o obj/Release/src/analysis.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/basicTypes.cpp -o obj/Release/src/basicTypes.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/createBarrier.cpp -o obj/Release/src/createBarrier.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/endOfGeneration.cpp -o obj/Release/src/endOfGeneration.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/endOfSimStep.cpp -o obj/Release/src/endOfSimStep.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/executeActions.cpp -o obj/Release/src/executeActions.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/feedForward.cpp -o obj/Release/src/feedForward.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/genome-compare.cpp -o obj/Release/src/genome-compare.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/genome.cpp -o obj/Release/src/genome.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/getSensor.cpp -o obj/Release/src/getSensor.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/grid.cpp -o obj/Release/src/grid.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/imageWriter.cpp -o obj/Release/src/imageWriter.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/indiv.cpp -o obj/Release/src/indiv.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/main.cpp -o obj/Release/src/main.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/params.cpp -o obj/Release/src/params.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/peeps.cpp -o obj/Release/src/peeps.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/random.cpp -o obj/Release/src/random.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/signals.cpp -o obj/Release/src/signals.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/simulator.cpp -o obj/Release/src/simulator.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/spawnNewGeneration.cpp -o obj/Release/src/spawnNewGeneration.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/survival-criteria.cpp -o obj/Release/src/survival-criteria.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/unitTestBasicTypes.cpp -o obj/Release/src/unitTestBasicTypes.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/unitTestConnectNeuralNetWiringFromGenome.cpp -o obj/Release/src/unitTestConnectNeuralNetWiringFromGenome.o
g++ -Wall -fexceptions -fopenmp -O3 -I/usr/include/opencv4 -c /home/dm/sw/biosim4-git/src/unitTestGridVisitNeighborhood.cpp -o obj/Release/src/unitTestGridVisitNeighborhood.o
g++ -o bin/Release/biosim4 obj/Release/src/analysis.o obj/Release/src/basicTypes.o obj/Release/src/createBarrier.o obj/Release/src/endOfGeneration.o obj/Release/src/endOfSimStep.o obj/Release/src/executeActions.o obj/Release/src/feedForward.o obj/Release/src/genome-compare.o obj/Release/src/genome.o obj/Release/src/getSensor.o obj/Release/src/grid.o obj/Release/src/imageWriter.o obj/Release/src/indiv.o obj/Release/src/main.o obj/Release/src/params.o obj/Release/src/peeps.o obj/Release/src/random.o obj/Release/src/signals.o obj/Release/src/simulator.o obj/Release/src/spawnNewGeneration.o obj/Release/src/survival-criteria.o obj/Release/src/unitTestBasicTypes.o obj/Release/src/unitTestConnectNeuralNetWiringFromGenome.o obj/Release/src/unitTestGridVisitNeighborhood.o -lX11 -lgomp -pthread -O3 -s /usr/lib/x86_64-linux-gnu/libopencv_core.so /usr/lib/x86_64-linux-gnu/libopencv_video.so /usr/lib/x86_64-linux-gnu/libopencv_videoio.so
Output file is bin/Release/biosim4 with size 778.42 KB
Process terminated with status 0 (0 minute(s), 11 second(s))
0 error(s), 0 warning(s) (0 minute(s), 11 second(s))