WorkflowHunt is a search engine for scientific workflow repositories.
This project has two important parts: semantic annotations and semantic search based on those semantic annotations.
We perform semantic annotations of workflows following the code below. First, we iterate over the ontologies available in the system. Second, we iterate over the terms of each ontology (also known as ontology classes). Third, we iterate over the workflows available in the system. Fourth, we verify if the ontology term is contained in the workflow metadata using exact string comparison if so, then we perform a semantic annotation using that ontology term and the workflow. Fifth, we verify if the synonym of the ontology term is contained in the workflow metadata if so, then we perform a semantic annotation using the ontology term associated with the synonym and the workflow.
A semantic annotation is a tuple (s, p, o, c), where s is the subject of annotation (workflow), o is the object of annotation (a term in a domain ontology), p is the predicate (the relationship between s and p), and c is the context (provenance information) [1].
def semantic_annotation()
foreach ontology in ontologies
foreach term in ontology
foreach workflow in workflows
if( term ⊆ workflow->metadata )
save( workflow, contains, term, { author, date } )
foreach synonym in term
if( synonym ⊆ workflow->metadata )
save( workflow, contains, term, { author, date } )
The semantic search algorithm uses a two-step approach:
Step 1. First, we iterate over the ontologies available in the system. Second, we iterate over the terms of each ontology. Third, we verify if the ontology term is contained in the user query using exact string comparison if so, then we store the ontology terms detected in the user query.
Step 2. First, we iterate over the semantic annotations stored in the system. Second, we select the workflows that have semantic annotations which match with the ontology term detected in Step 1. Finally, we return those workflows (without ranking their relevance).
def semantic_search( query )
terms_detected = ∅
results = ∅
// First Step: Detecting ontology terms in the query
foreach ontology in ontologies
foreach term in ontology
if( term ⊆ query )
terms_detected.add( term )
foreach synonym in term
if( synonym ⊆ workflow->metadata )
terms_detected.add( term )
// Second Step: Searching workflows that have semantic annotations with
// ontology terms in terms_detected
foreach sa in semantic_annotations
if( sa->o ⊆ terms_detected )
workflow = search_workflow( sa->o )
results.add( workflow )
return results
For this project, we used the ontologies EDAM and CHEMINF. EDAM is an ontology of bioinformatics information, including operations, types of data, topics, and formats [2]. CHEMINF is an ontology of chemical information, including terms, and algorithms used in chemistry [3]. The ontology terms and synonyms were collected from the Ontology Lookup Service. Ontology Lookup Service is a service that allows querying, browsing, and navigating over a database that integrates several biomedical ontologies and related vocabulary [4]. Moreover, we use WordNet [5] as an additional source of synonyms to complement the synonyms collected in the Ontology Lookup Service.
We use an exact string comparison between the text in the workflow metadata and the ontology terms and synonyms to produce semantic annotations. Nevertheless, we set the restriction of adding a white space at the start and end of each term and synonym to decrease the possibilities of false positives in the semantic annotations. For example, consider the word "ph" considered a synonym of the ontology term "PHYLIP format" in the EDAM ontology. Without those spaces is possible to perform wrong semantic annotations for texts like "Phylogenetic" that can be annotated with the ontology term "PHYLIP Format" just because "ph" is a substring of "Phylogenetic".
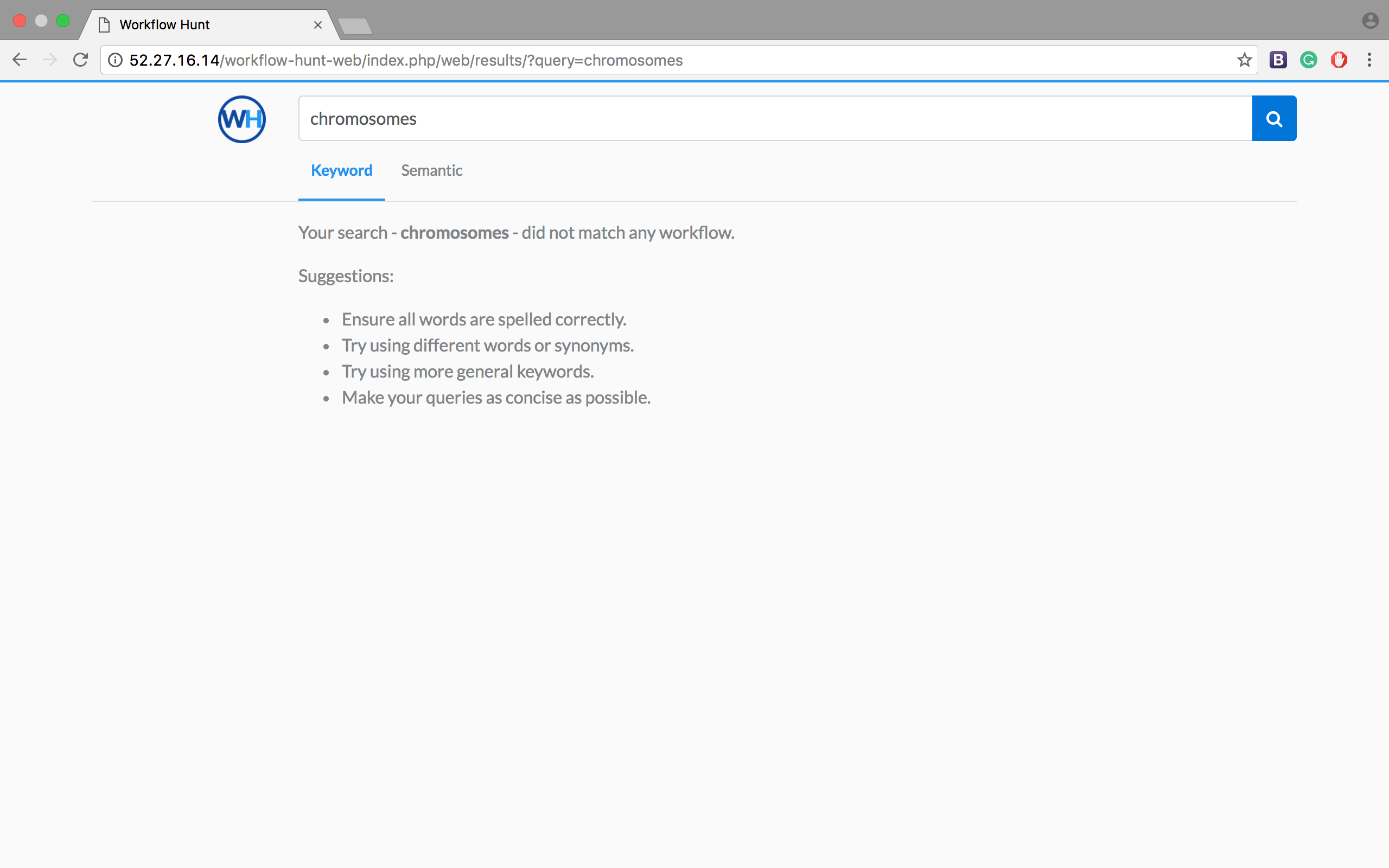
The results of this project are available here. We can see the difference between keyword-based search and semantic-based search with a query like "chromosomes".
That word is contained in the metadata of just one workflow. Thus, the keyword-based search should show just that result. Nevertheless, as we can see in Figure 1, our current implementation of the keyword-based search in ElasticSearch judges that result with a low score and does not return it.
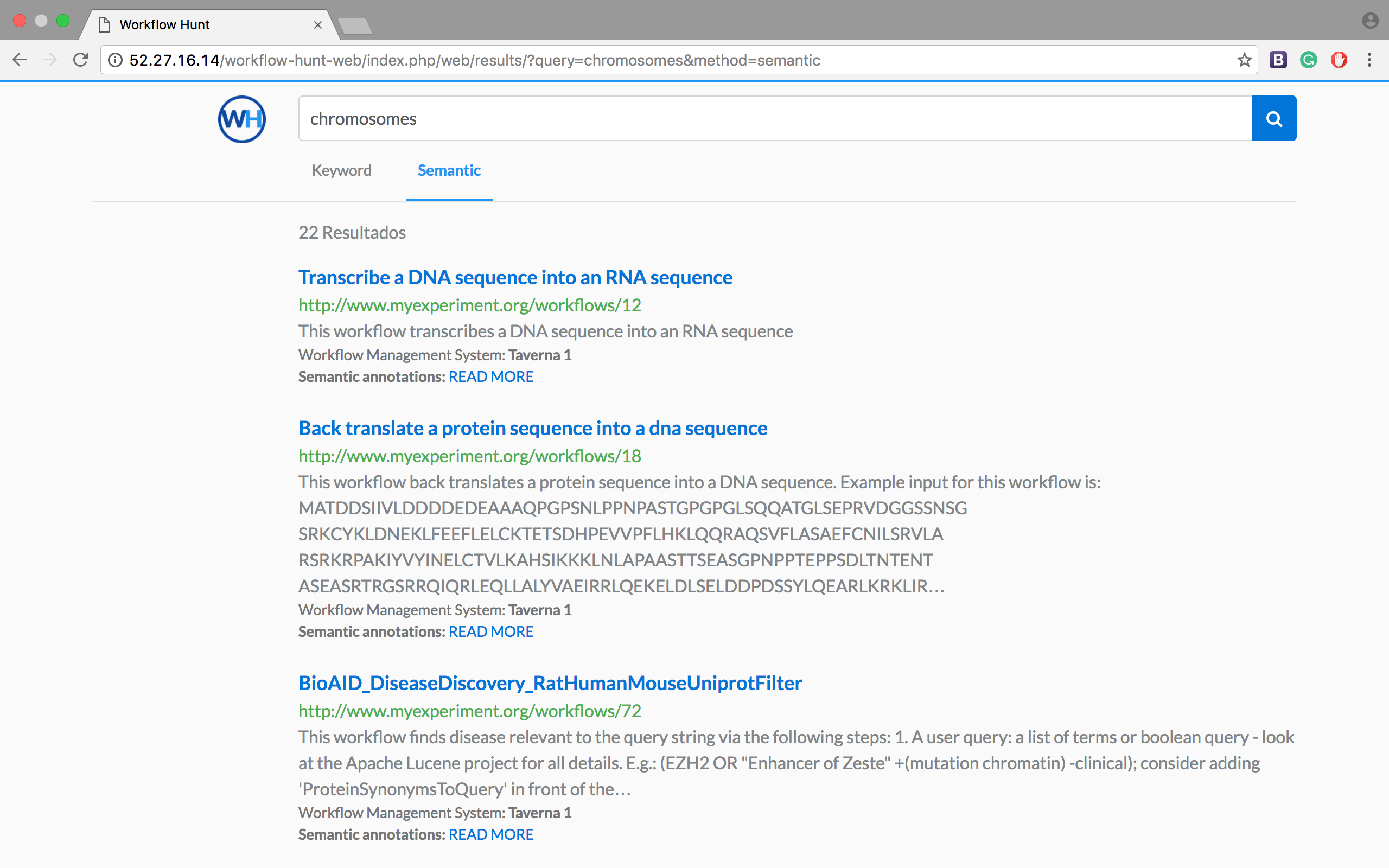
On the other hand, in Figure 2 we can see that the semantic-based search returns 22 results associated with that query (without ranking their relevance). These results are shown because according to the EDAM ontology, the word "chromosomes" is semantically related to terms like "Ancient DNA", "DNA Analysis" and "DNA".
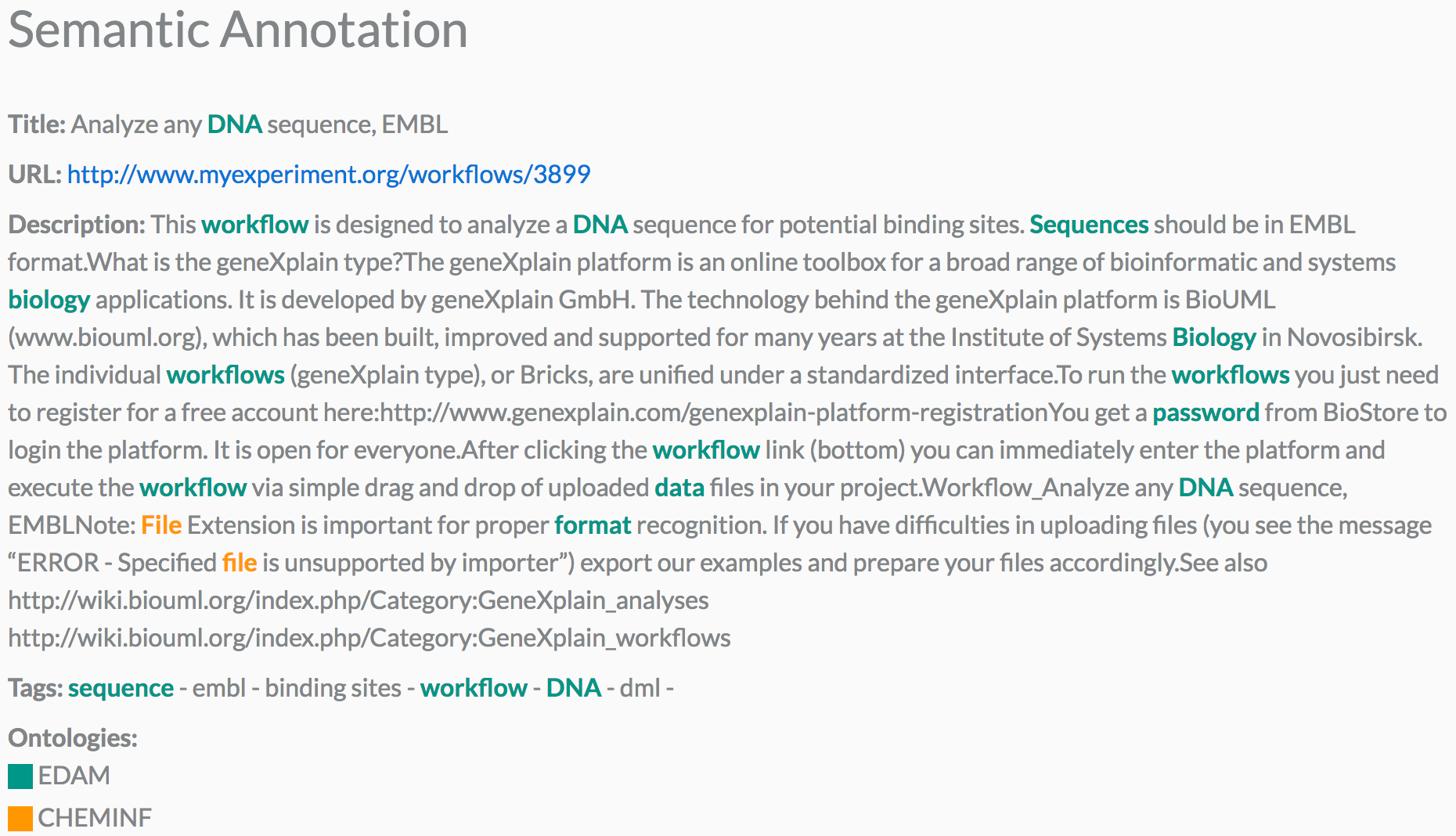
Moreover, we can see in Figure 3 a graphical representation of the semantic annotations in the workflow metadata. This graphical representation is available in the retrieval system by clicking on "READ MORE", which is present in all the semantic results.
[1] Oren, E., Möller, K., Scerri, S., Handschuh, S., & Sintek, M. (2006). What are semantic annotations. Technical report. DERI Galway, 9, 62.
[2] Ison, J., Kalaš, M., Jonassen, I., Bolser, D., Uludag, M., McWilliam, H., ... & Rice, P. (2013). EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics, 29(10), 1325-1332.
[3] Hastings, J., Chepelev, L., Willighagen, E., Adams, N., Steinbeck, C., & Dumontier, M. (2011). The chemical information ontology: provenance and disambiguation for chemical data on the biological semantic web. PloS one, 6(10), e25513.
[4] Côté, R. G., Jones, P., Apweiler, R., & Hermjakob, H. (2006). The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC bioinformatics, 7(1), 97.
[5] Christiane Fellbaum (1998, ed.) WordNet: An Electronic Lexical Database. Cambridge, MA: MIT Press.