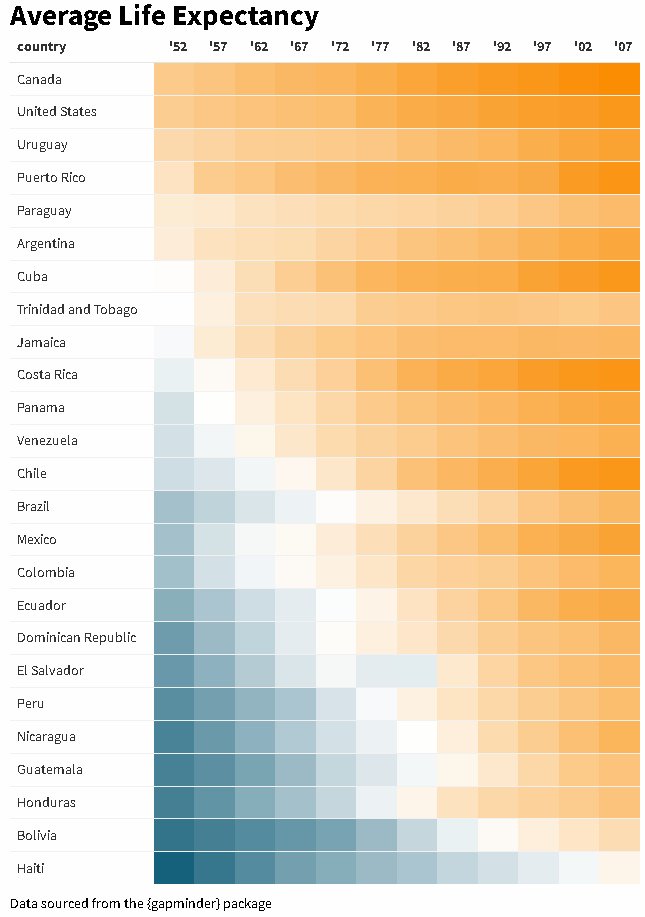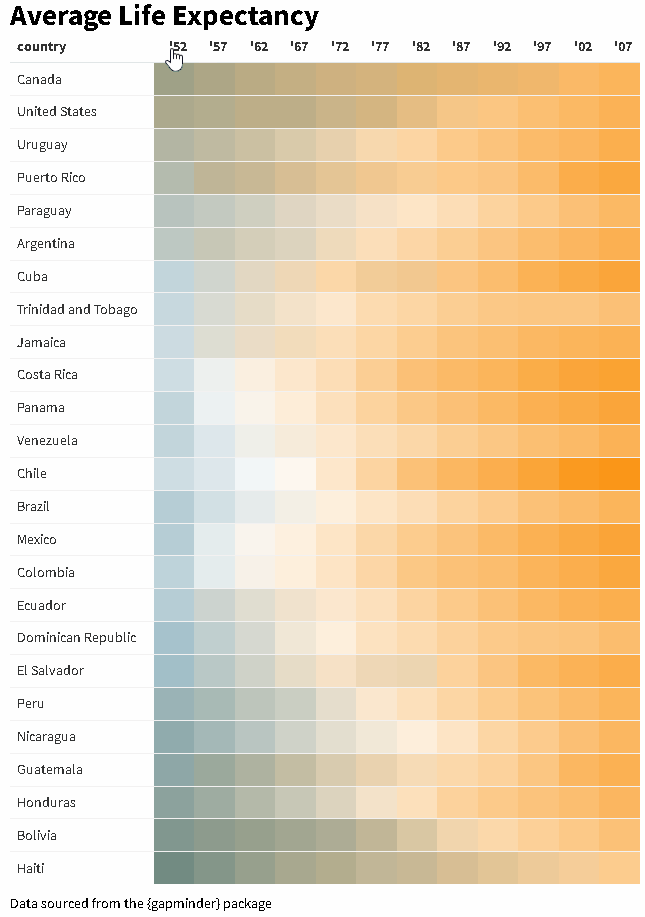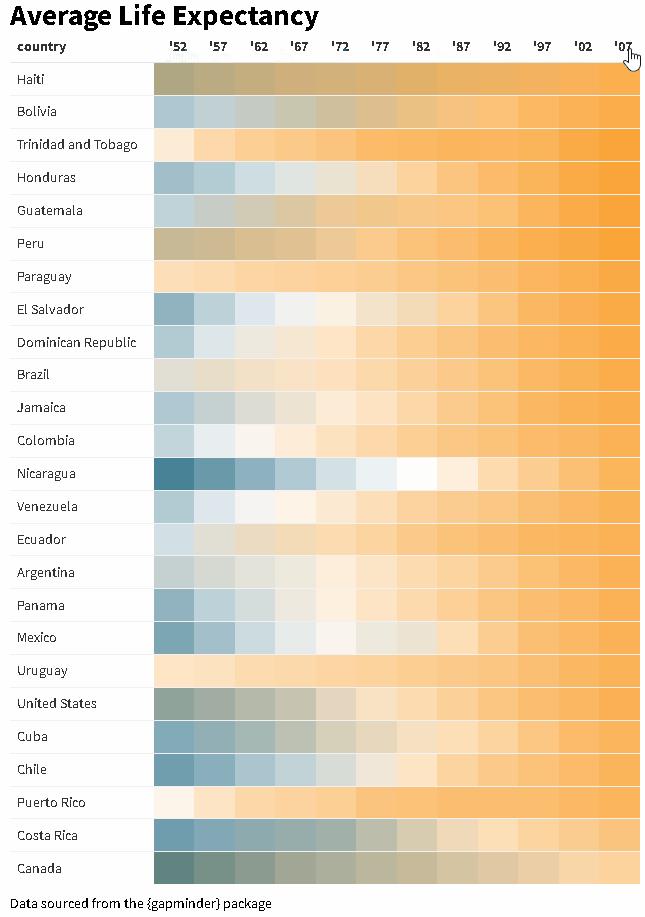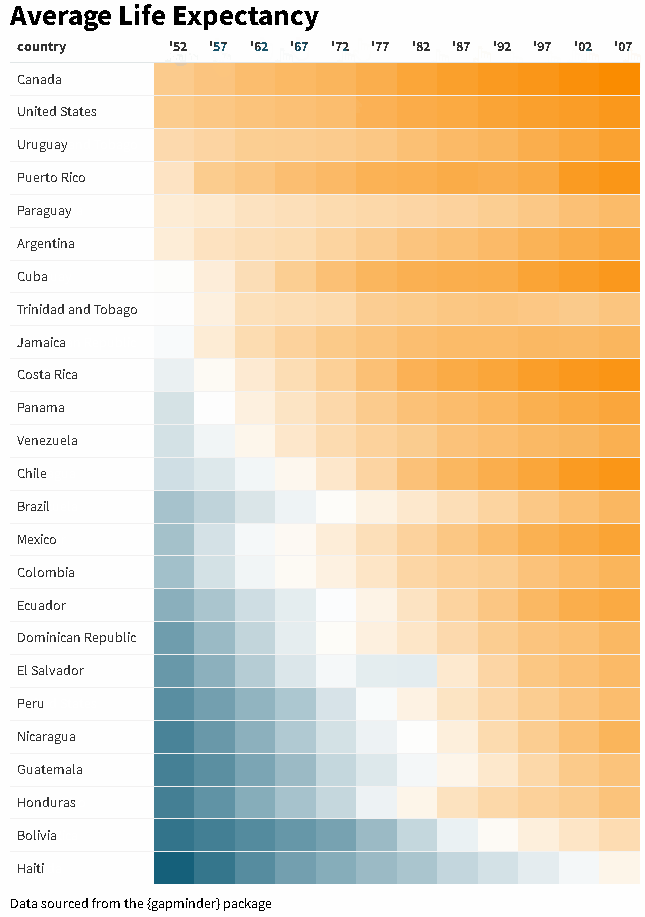
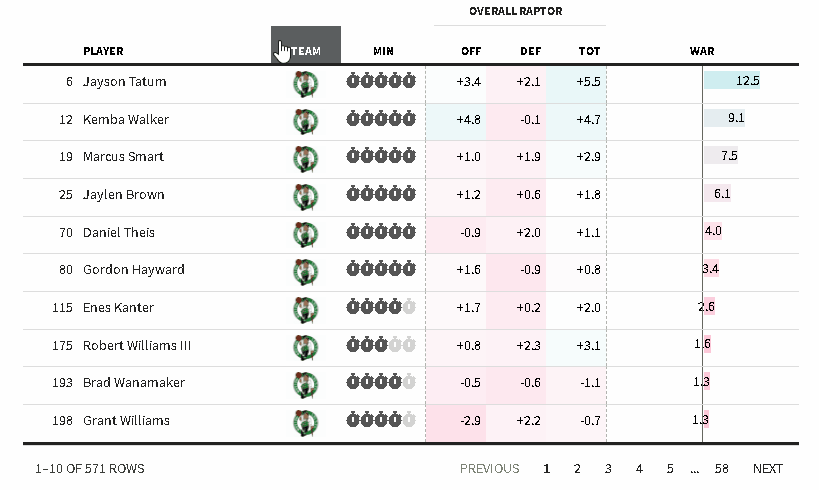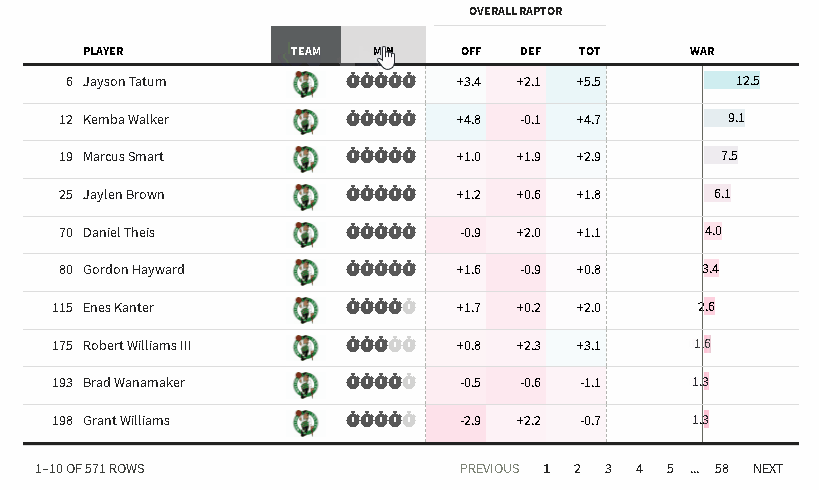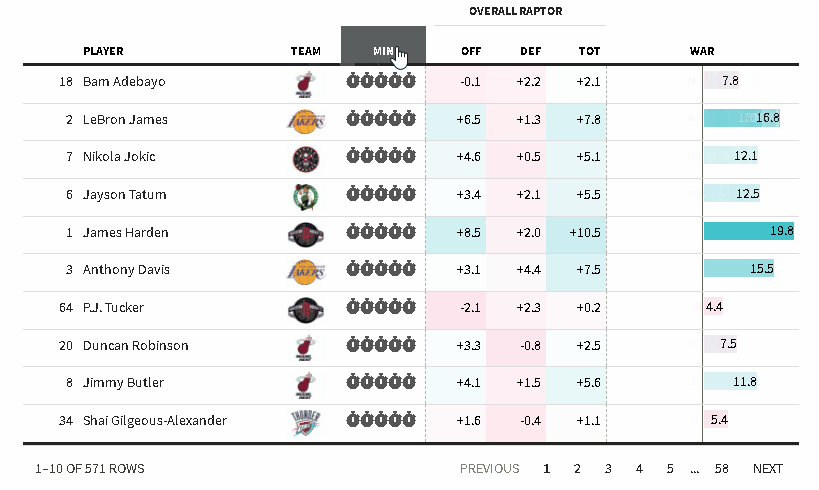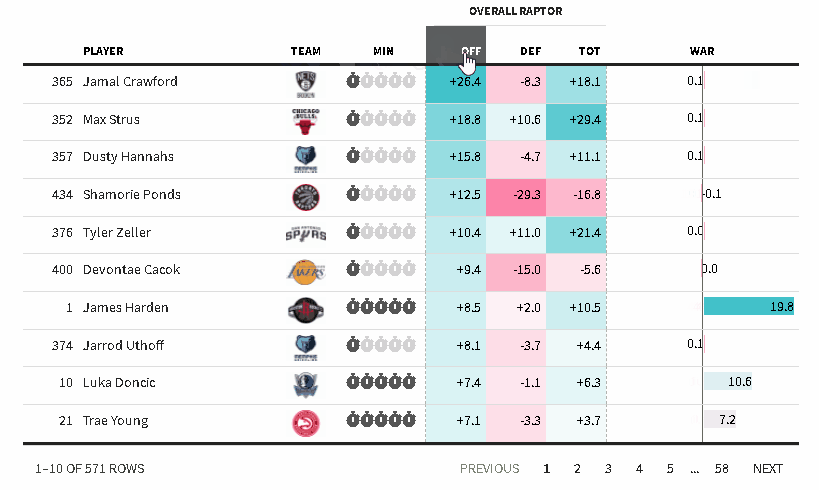
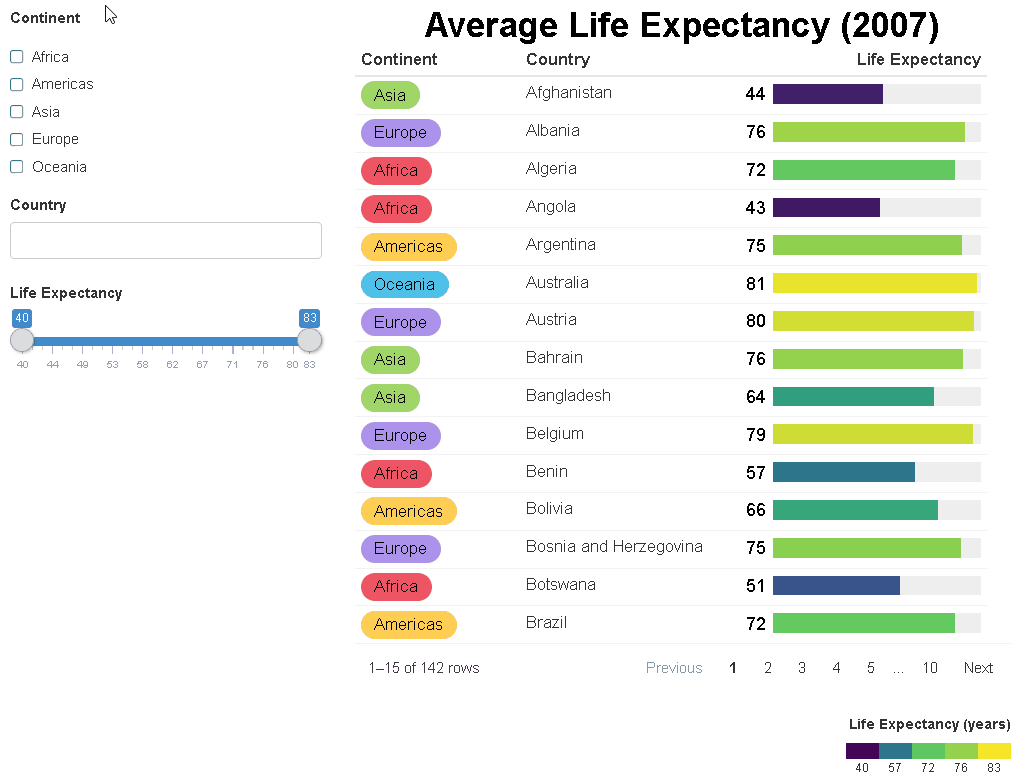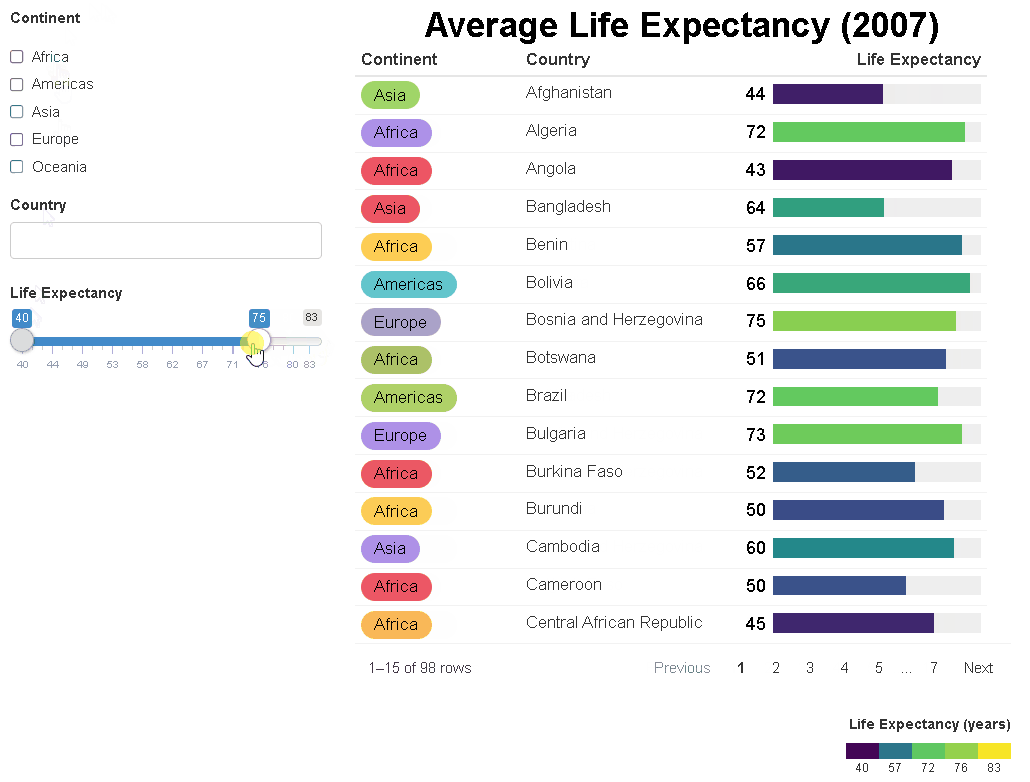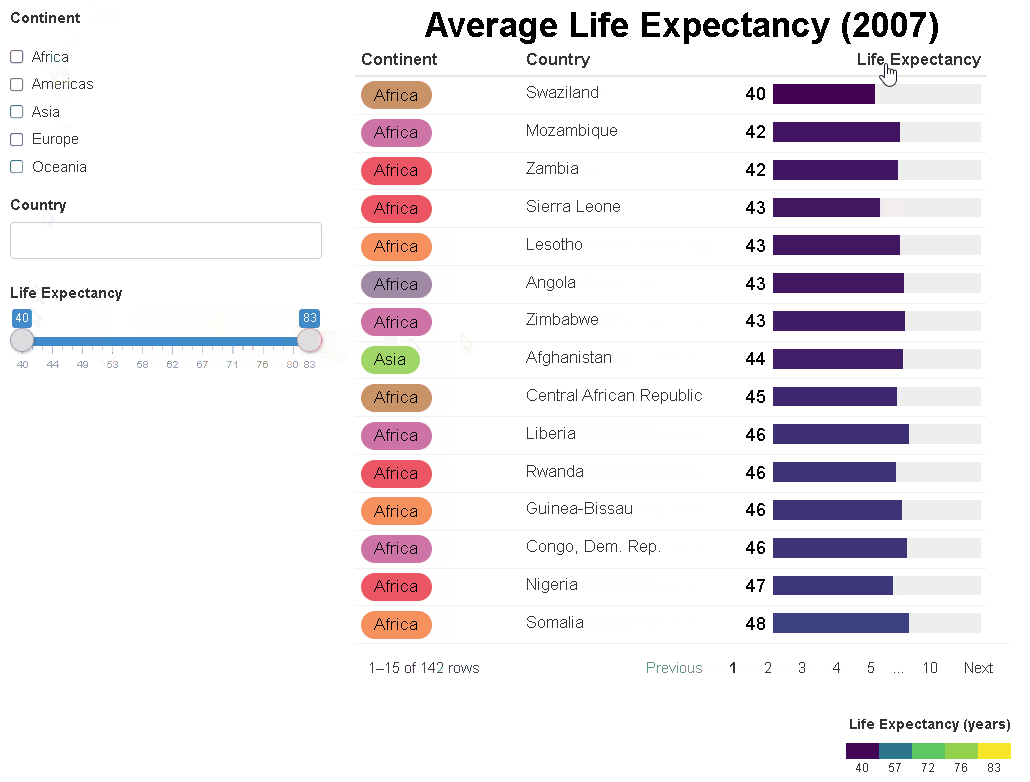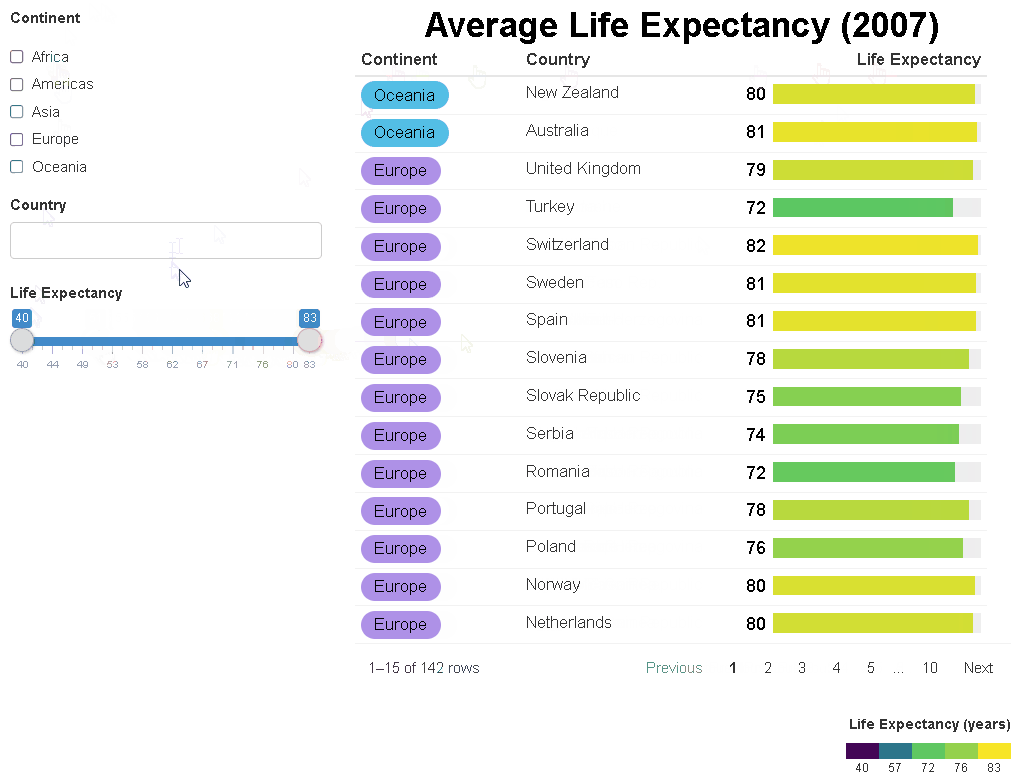
The {reactablefmtr} package streamlines and enhances the styling and formatting of tables built with the {reactable} R package. The {reactablefmtr} package provides many conditional formatters that are highly customizable and easy to use.
-
Conditionally format tables with color scales, color tiles, and data bars. Assign icons from Font Awesome with icon assign and icon sets.
-
Interactive sparklines that are highly customizable and available in line or bar format.
-
Custom table themes that can easily be applied to any {reactablefmtr} or {reactable} table.
-
Embed images directly from the web into your table.
-
Save tables as static PNG files or as interactive HTML files.
For more examples, check out the vignettes. To stay up to date with the latest upgrades to the development version, be sure to follow the package news.
The {reactablefmtr} package is available from CRAN and can be installed with:
install.packages("reactablefmtr")
Or install the development version of {reactablefmtr} with:
remotes::install_github("kcuilla/reactablefmtr")
The {reactable} package allows for the creation of interactive data tables in R. However, styling tables within {reactable} requires a lot of code, can be difficult for many R users (due to the need to understand HTML and CSS), and not scalable. For example, here is the current method of creating bar charts within {reactable}:
library(reactable)
library(htmltools)
data <- MASS::Cars93[20:49, c("Make", "MPG.city", "MPG.highway")]
bar_chart <- function(label, width = "100%", height = "16px", fill = "#15607A", background = "#EEEEEE") {
bar <- div(style = list(background = fill, width = width, height = height))
chart <- div(style = list(flexGrow = 1, marginLeft = "8px", background = background), bar)
div(style = list(display = "flex", alignItems = "center"), label, chart)
}
reactable(
data,
columns = list(
MPG.city = colDef(align = "left", cell = function(value) {
width <- paste0(value / max(data$MPG.city) * 100, "%")
bar_chart(value, width = width)
}),
MPG.highway = colDef(align = "left", cell = function(value) {
width <- paste0(value / max(data$MPG.highway) * 100, "%")
bar_chart(value, width = width)
})
)
)
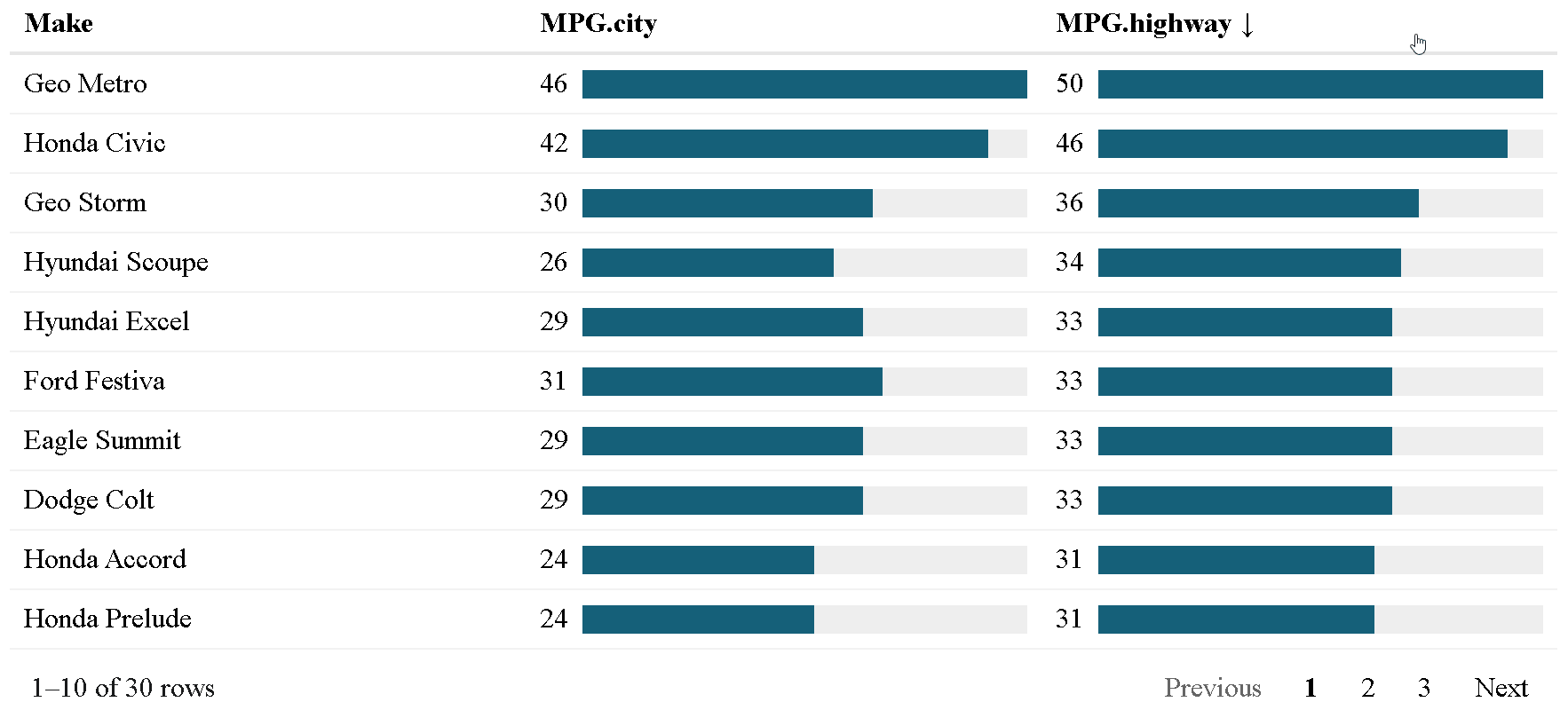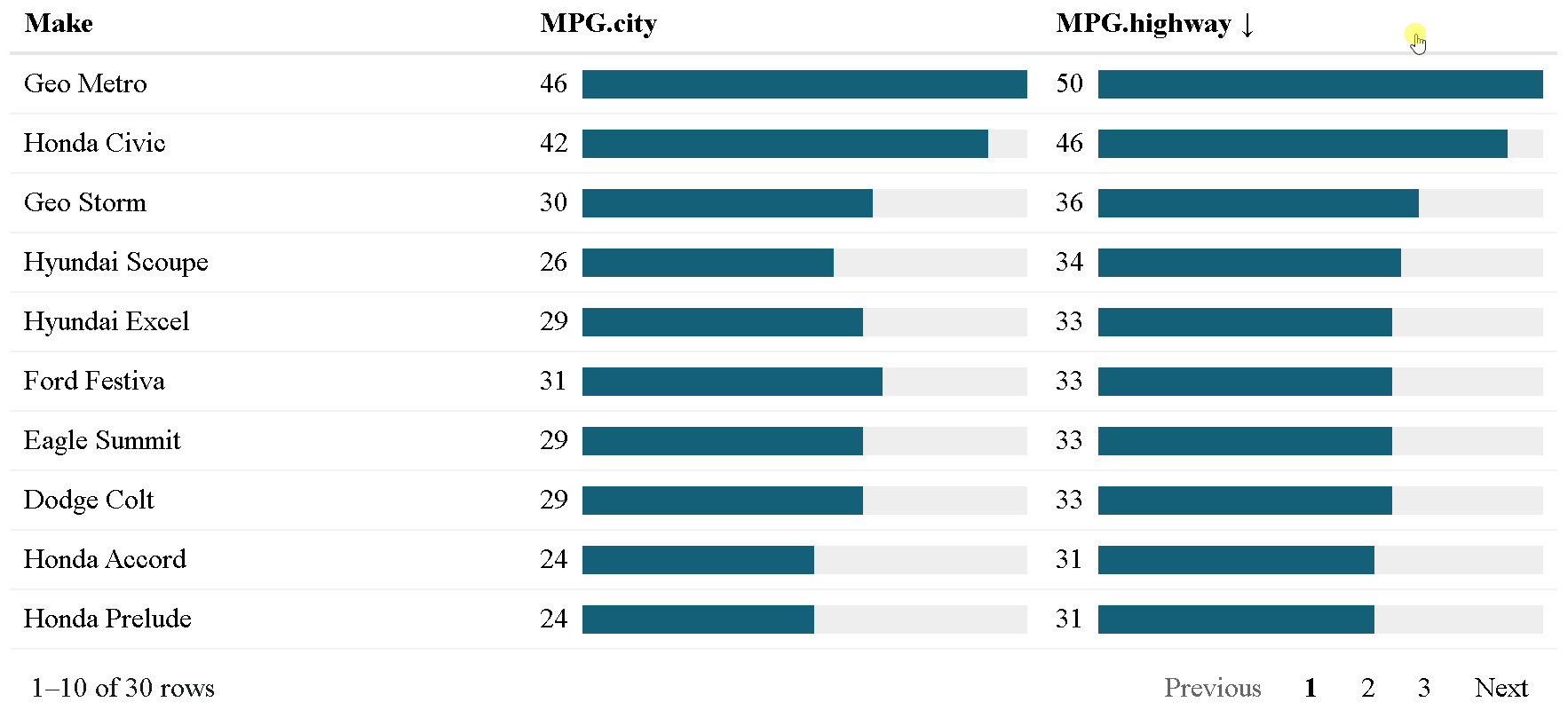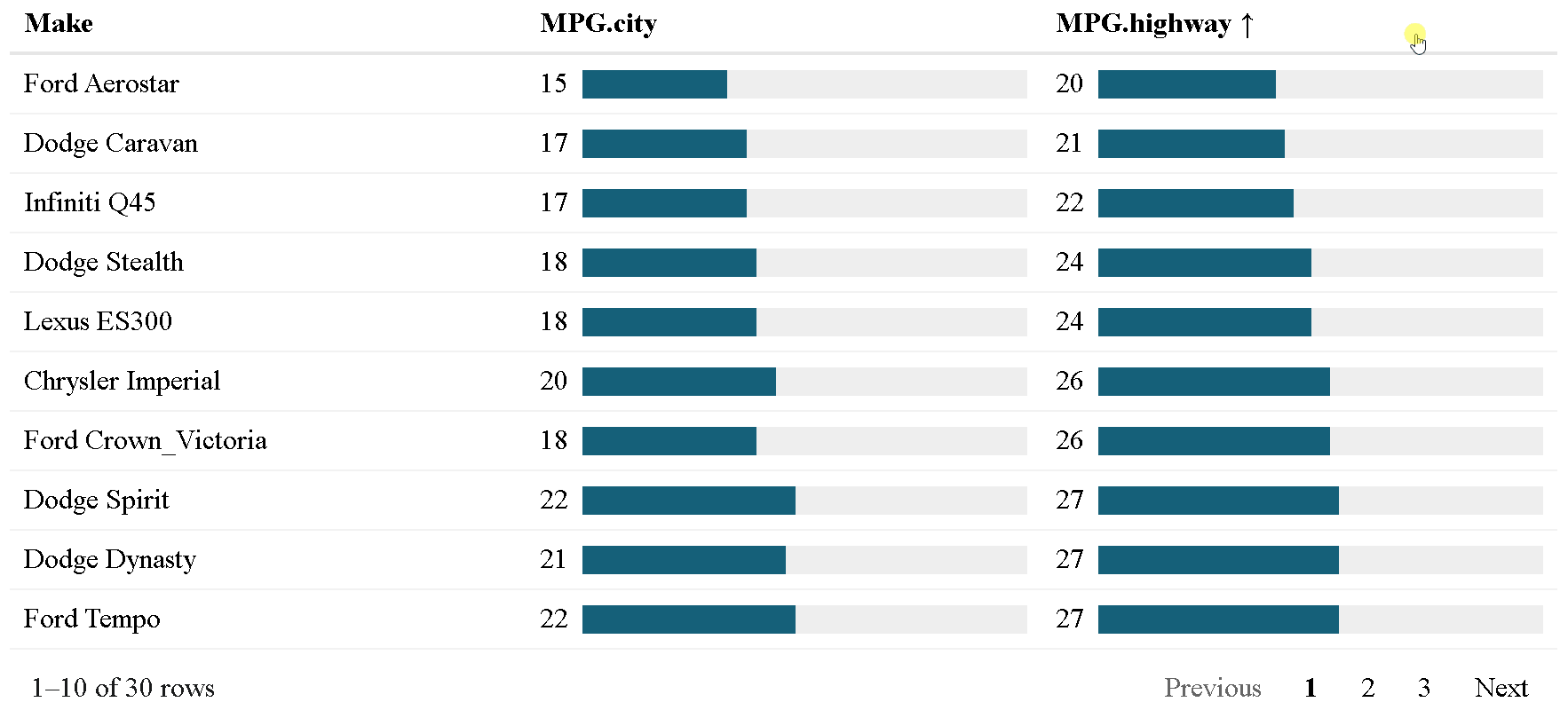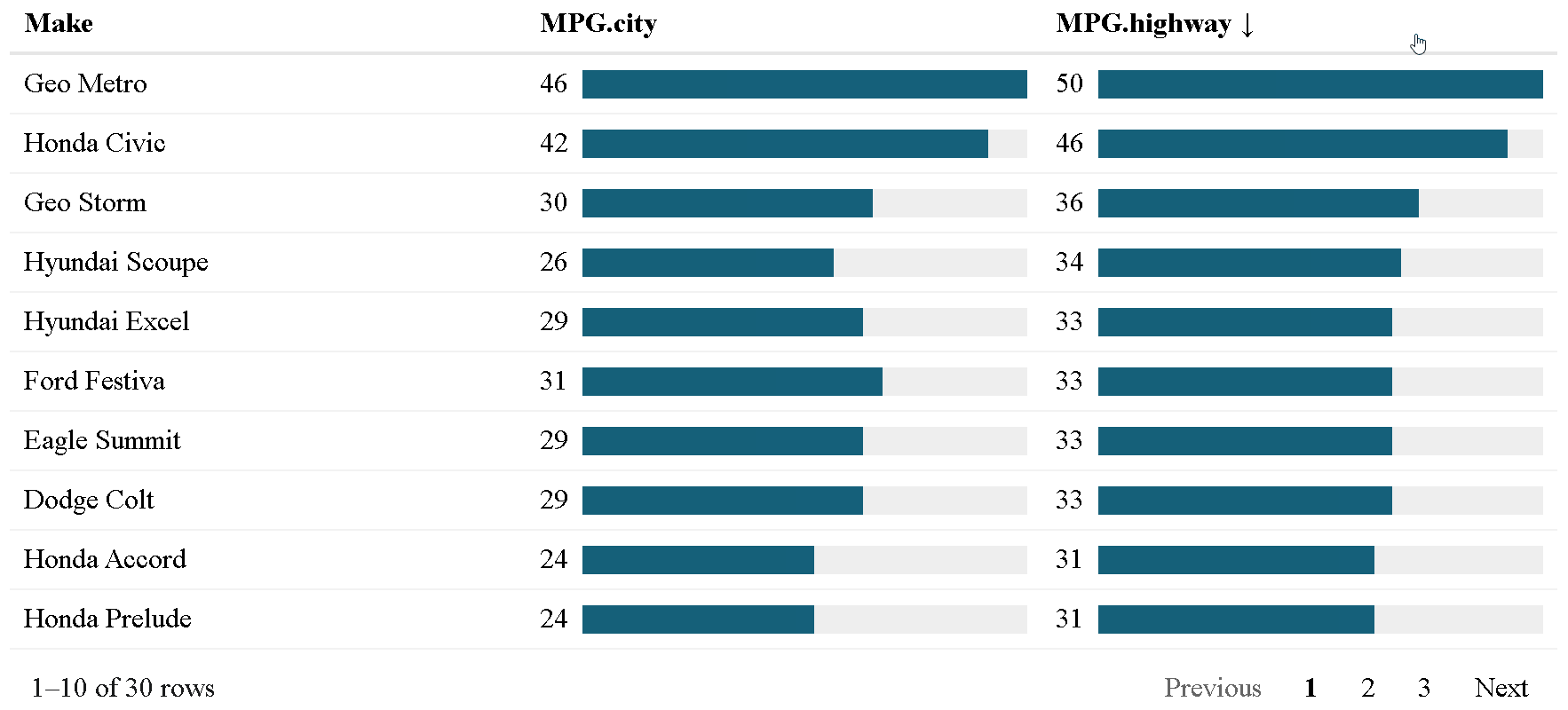
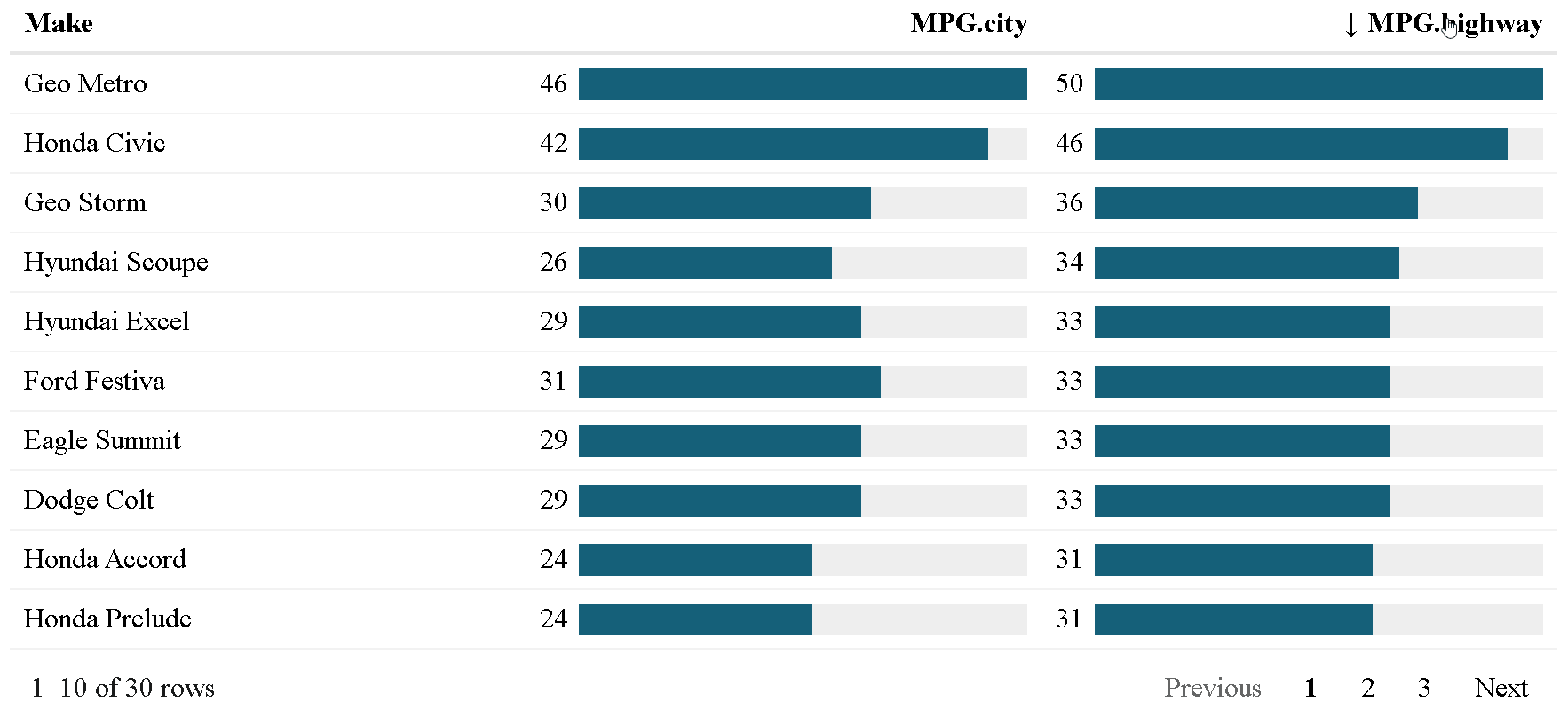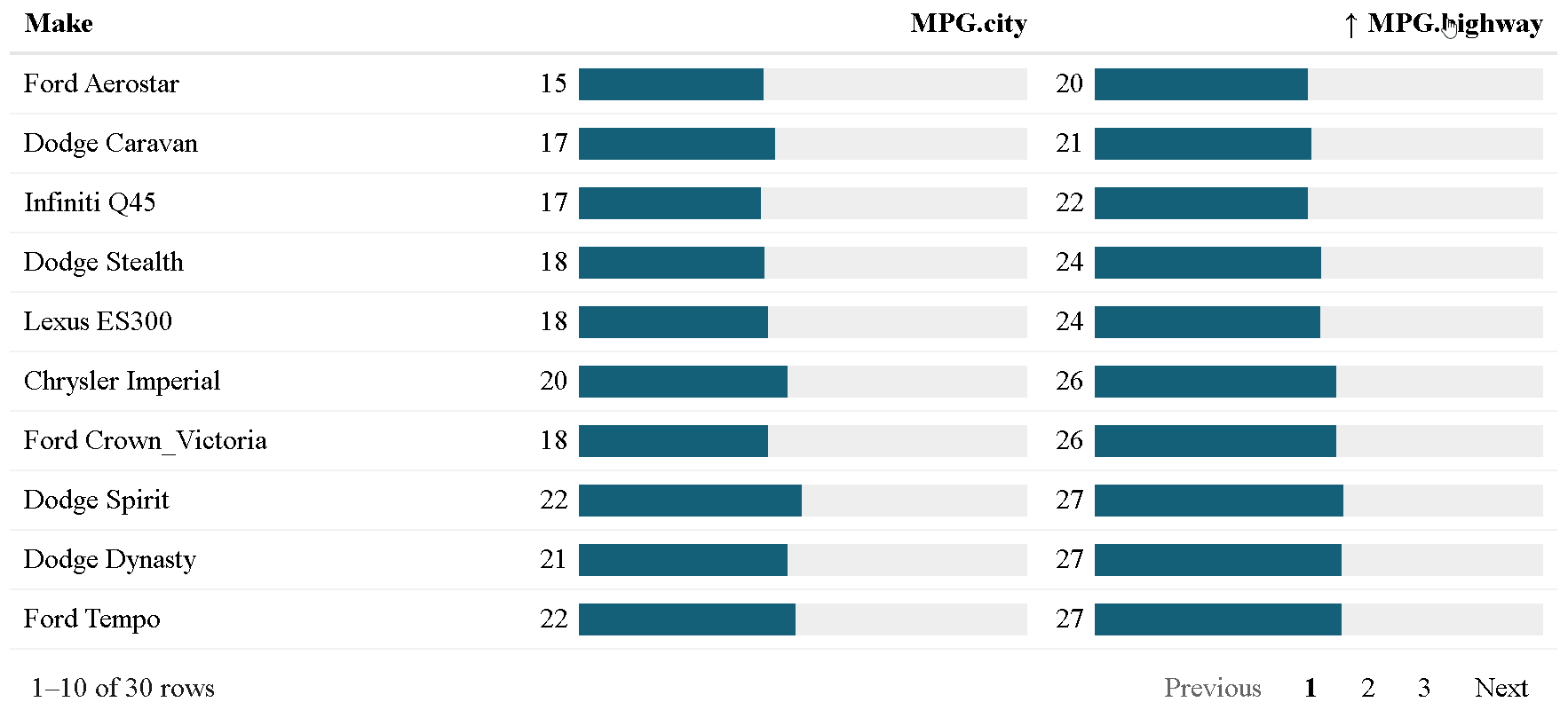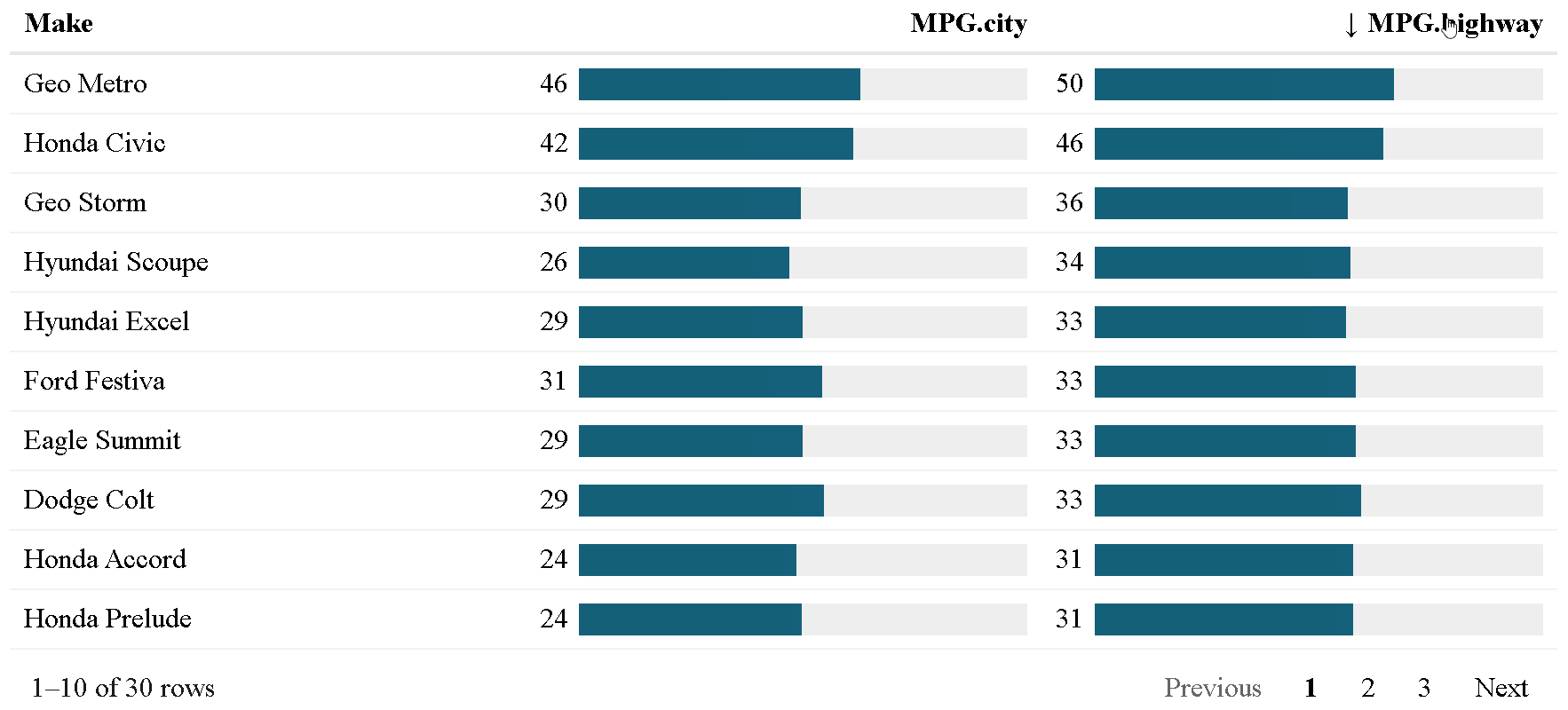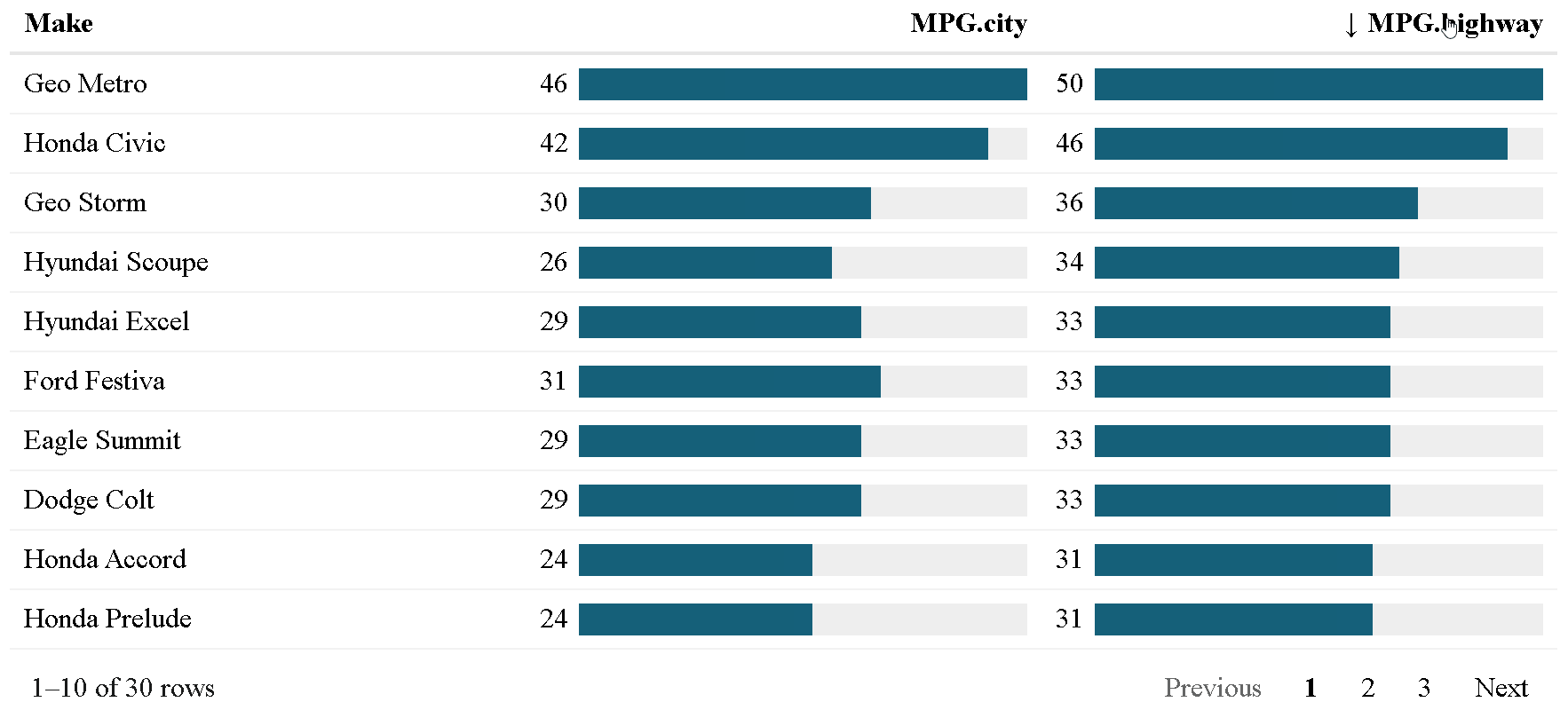
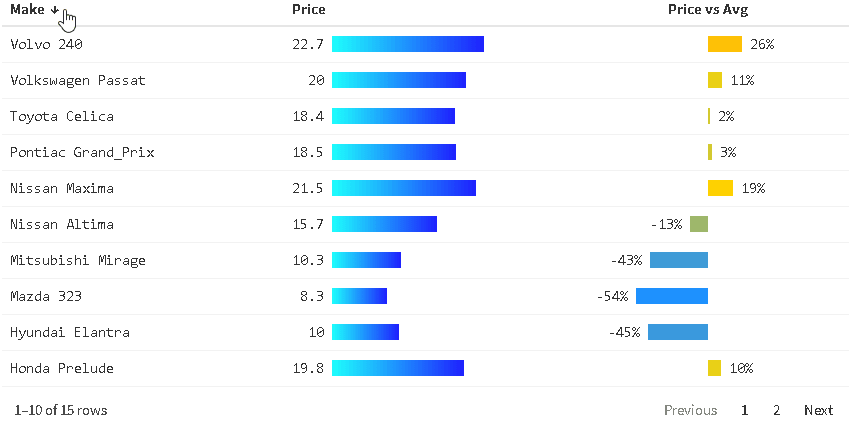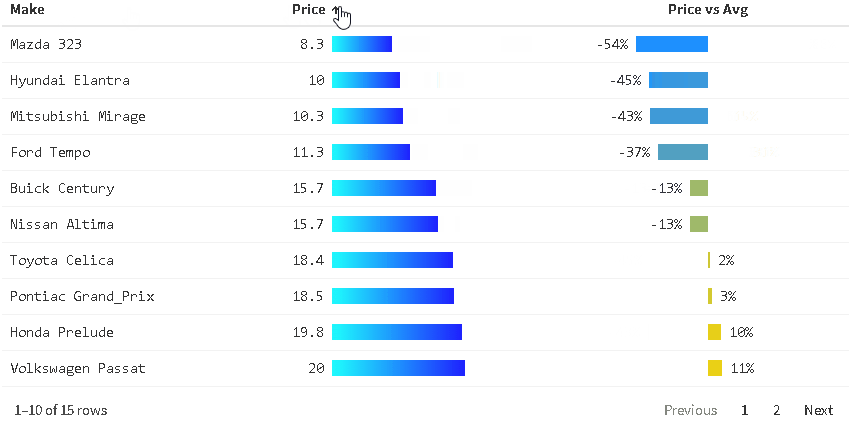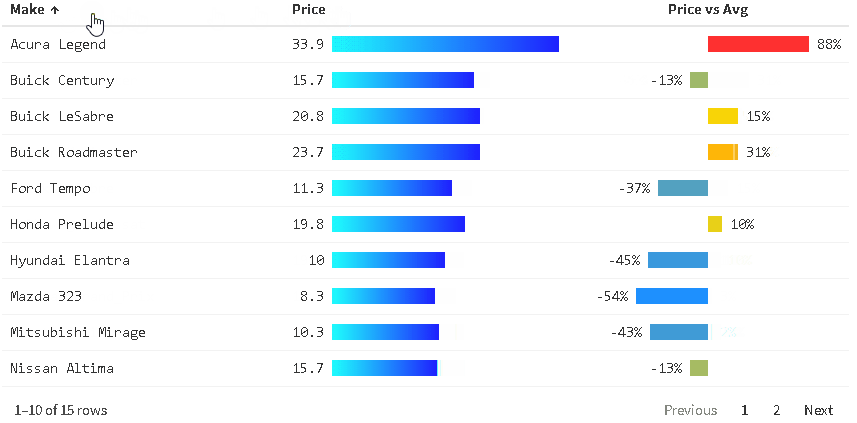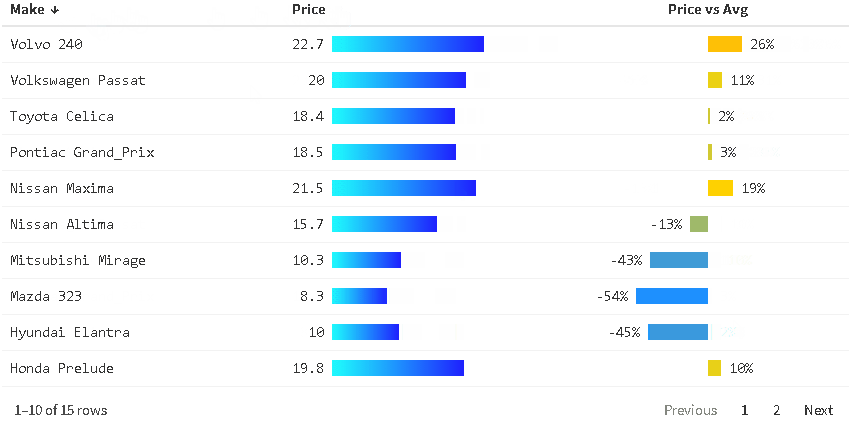
The {reactablefmtr} package presents a much simpler method of creating bar charts with the data_bars() function. In addition to needing far less code, there are also a multitude of customization options available to easily change the appearance of the bar charts, and as an added bonus, the bars animate on sort!
library(reactablefmtr)
reactable(
data,
defaultColDef = colDef(
cell = data_bars(data, text_position = "outside-base")
)
)
Use data_bars() to assign horizontal bars to each row. There are many ways to customize the look of data_bars(), including the alignment of the bars, the position of the text labels, and the option to add icons and images to the bars. See the tutorial for customization examples.
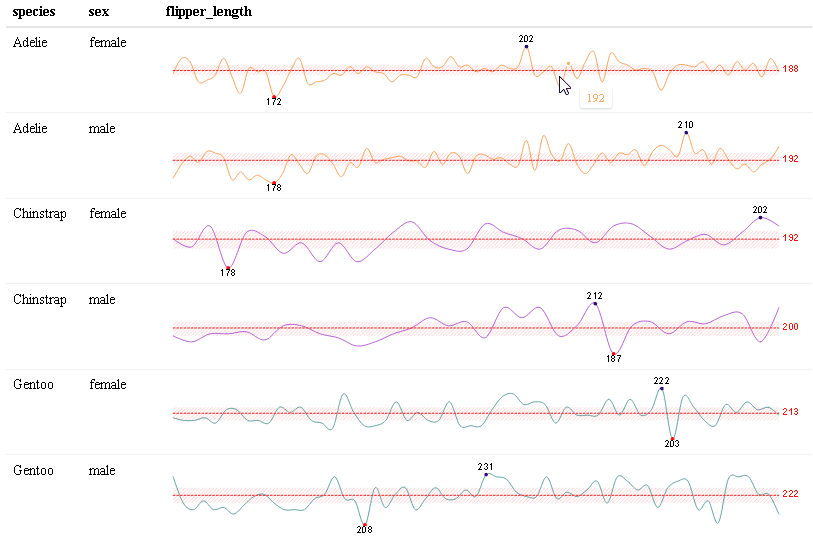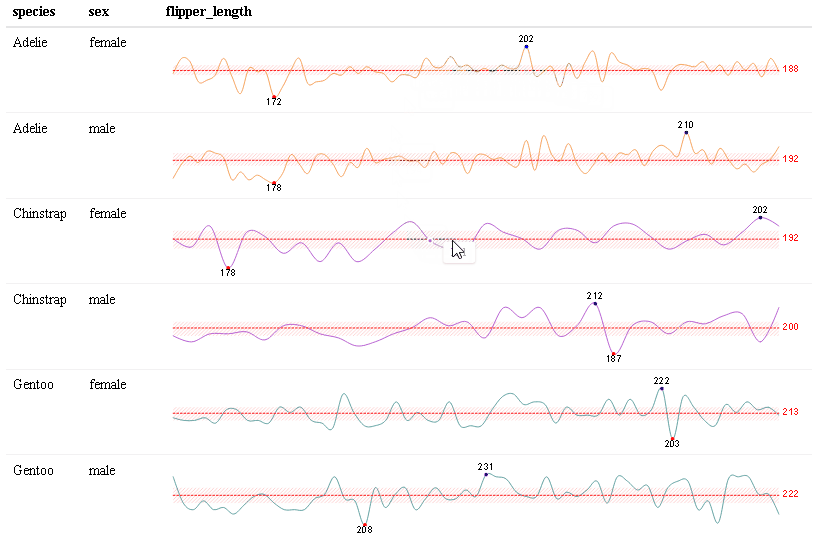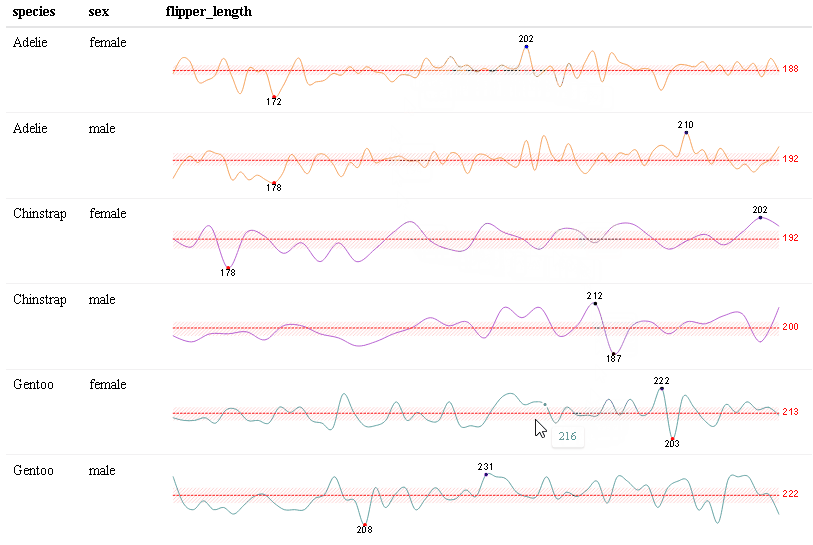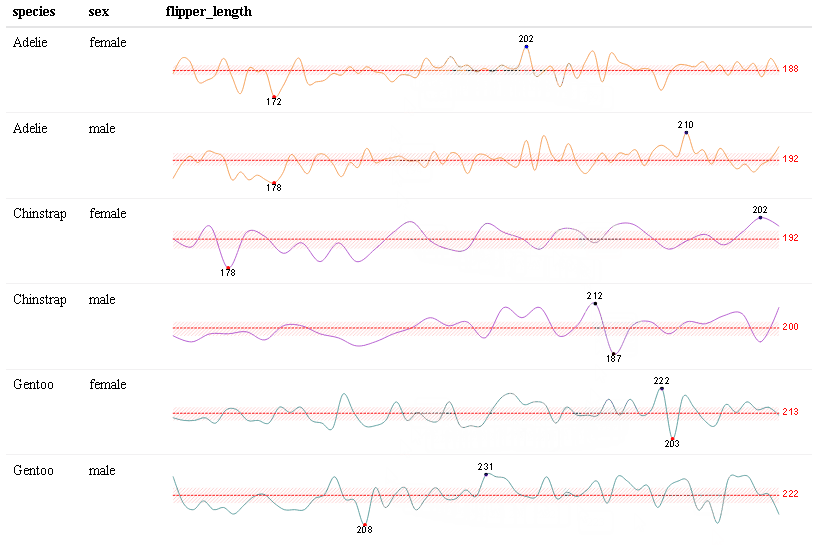
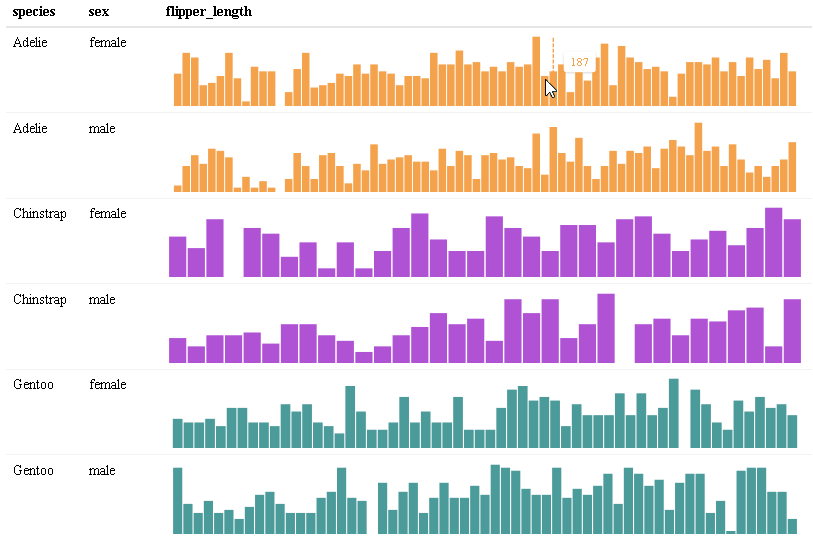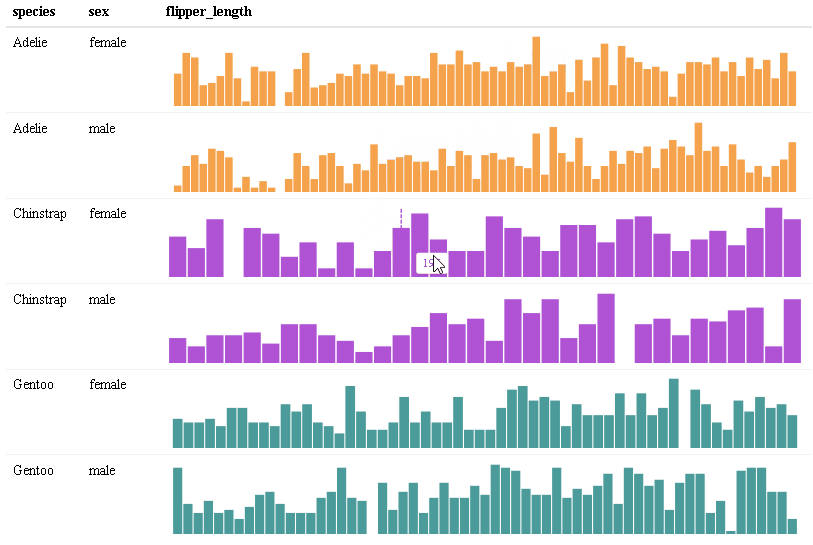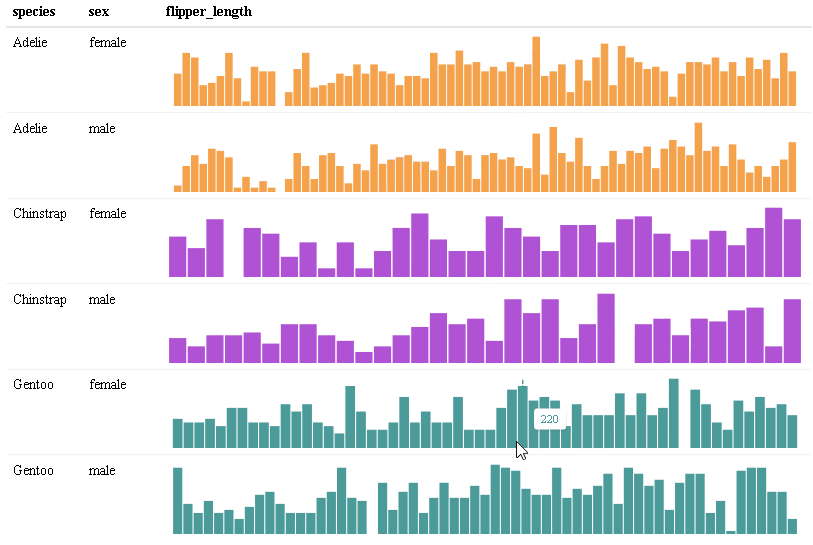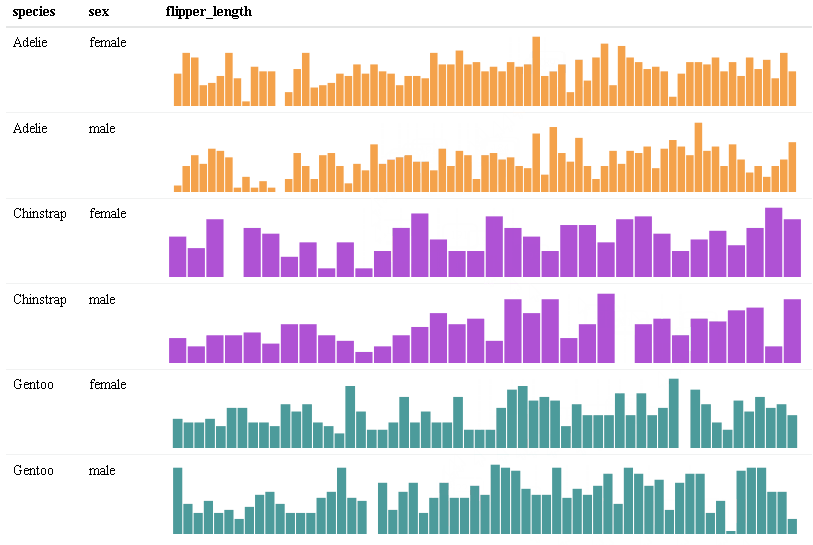
Use react_sparkline() to create sparklines or react_sparkbar() to create sparkline bar charts. The sparklines are highly customizable and interactive. As you hover over each data point, the value will appear. The sparklines are imported from the {dataui} package, so this package will need to be downloaded from GitHub in order to use this feature.
Use color_scales() to assign conditional colors to cells based on their relative values. The color of the text in the cells automatically adjusts based on the shade of the cell color, allowing the use of dark-colored palettes. See the tutorial for more examples.
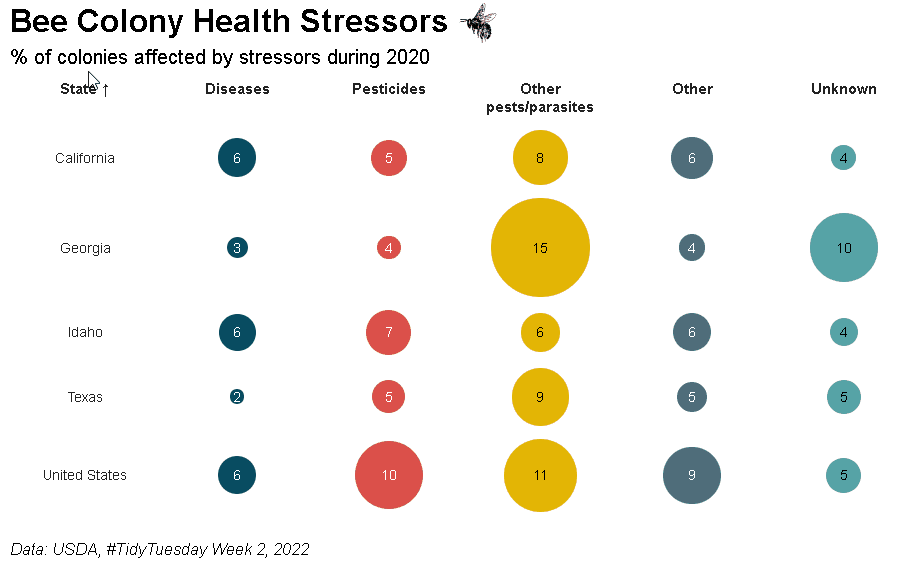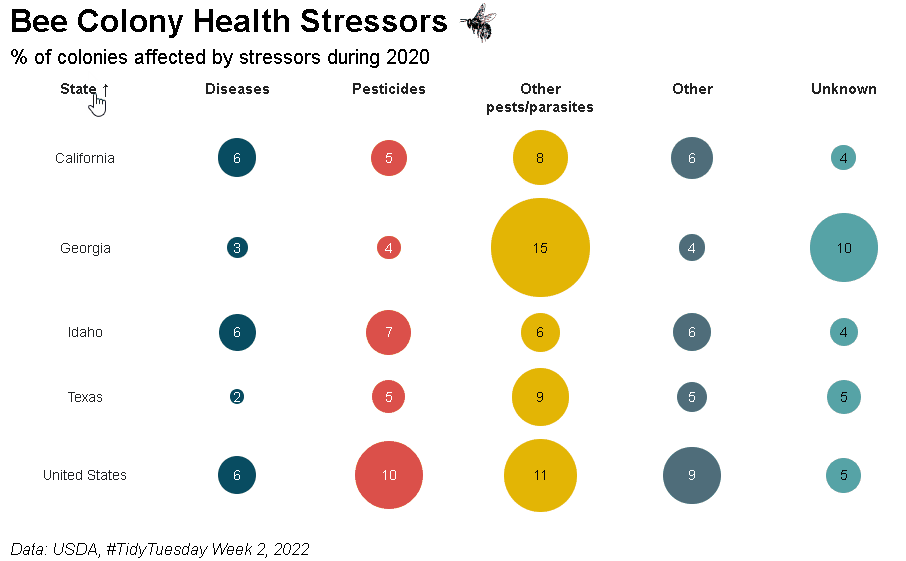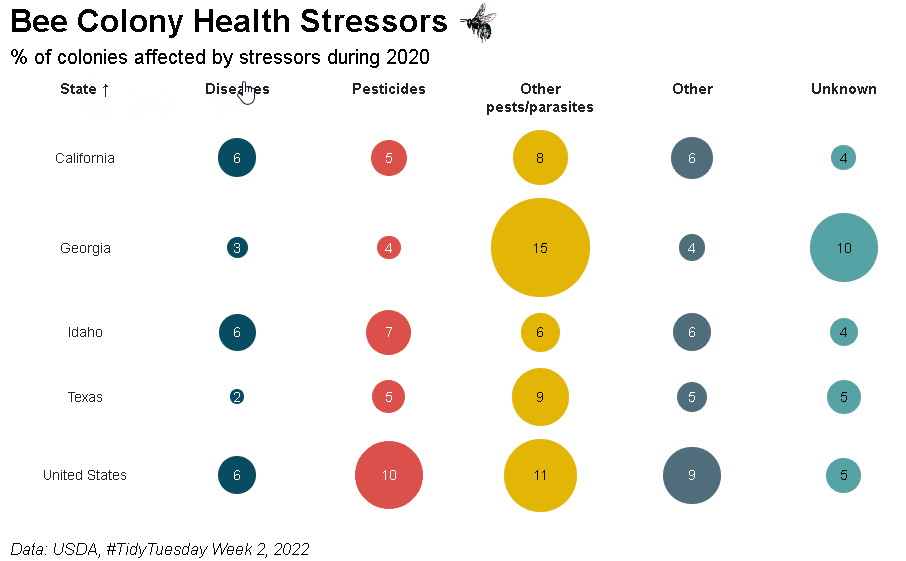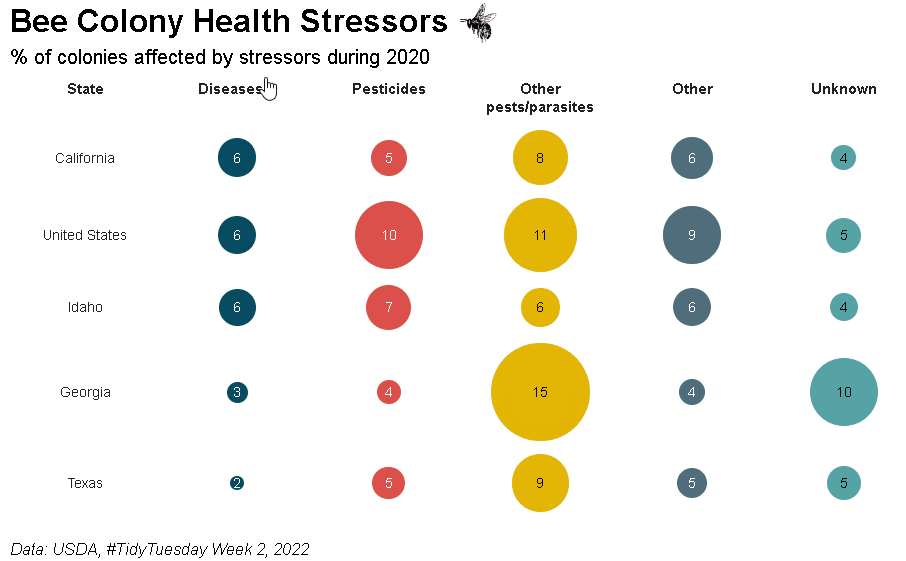
Bubble grid tables can be created using bubble_grid(). Bubbles can be shown as circles or squares using the shape parameter. To see how the example below was created, please check out the bubble grids section in the {reactablefmtr} cookbook:
Within {reactablefmtr}, there are 24+ custom table themes. The themes include bootstrap themes, themes inspired by news/sports sites such as The New York Times, FiveThirtyEight, and ESPN, as well as other custom themes that can only be found within {reactablefmtr}. The themes can be applied easily to tables by simply referencing the theme name. Additional customization options, such as changing the font size, font color, etc. are also available.
An example of the fivethirtyeight() theme:
{reactablefmtr} works well when linked to UI controls via {crosstalk} or within a Shiny app.
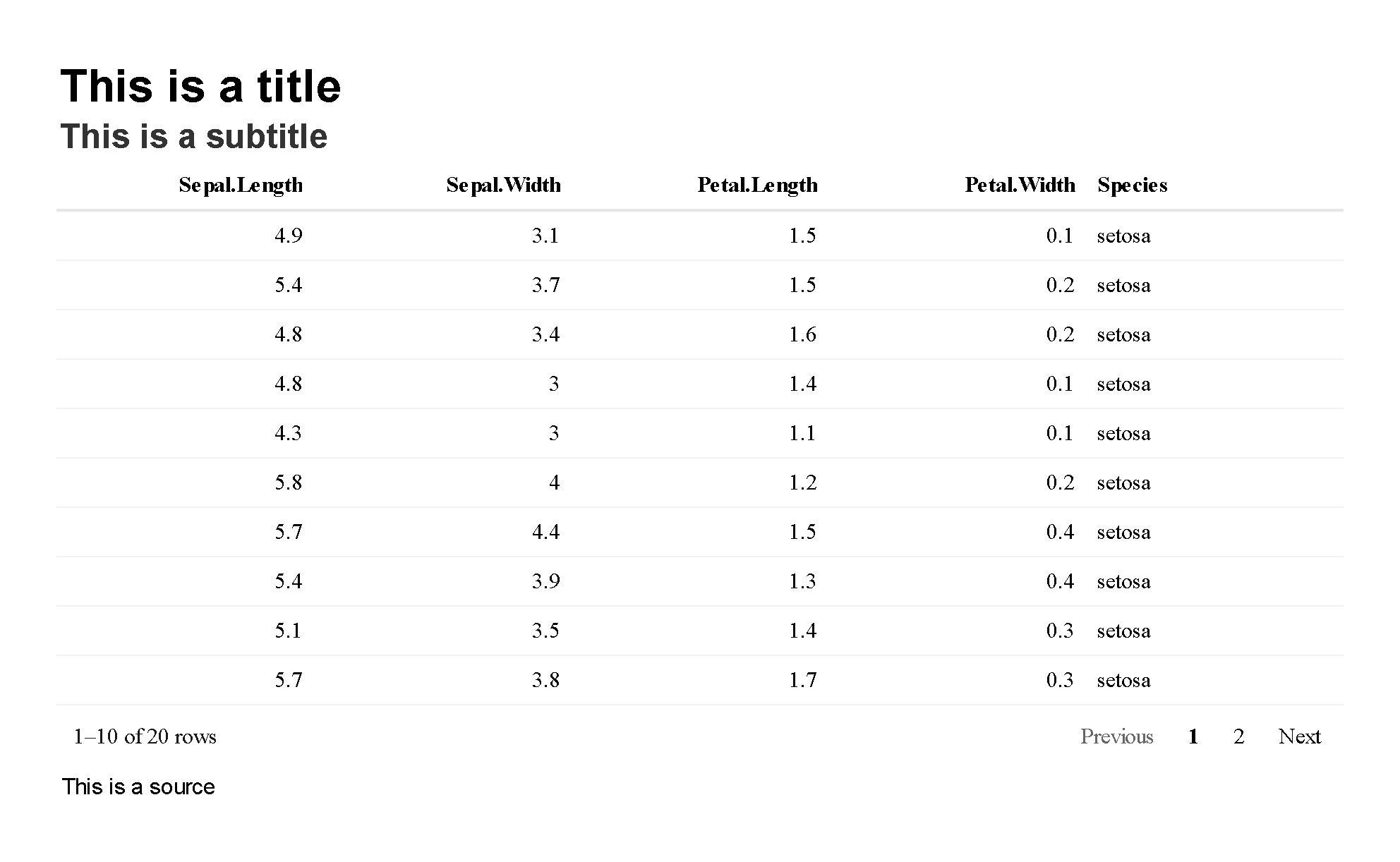
Titles and subtitles can be easily placed above any {reactablefmtr} or {reactable} table with add_title() and add_subtitle(). Also have the option to include a source below a table with add_source(). Additional customization options such as changing the alignment, font size, font family, font style, and font color are available within each formatter.
reactable(iris[10:29, ]) %>%
add_title("This is a title") %>%
add_subtitle("This is a subtitle") %>%
add_source("This is a source")
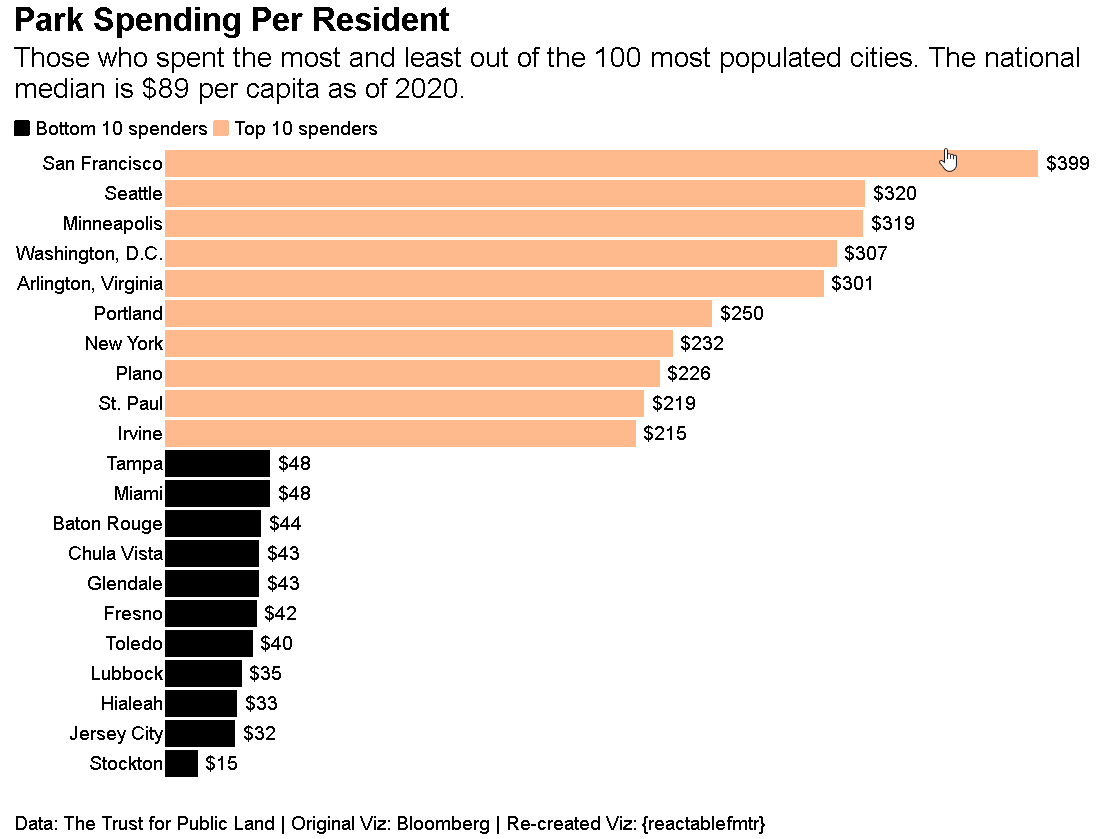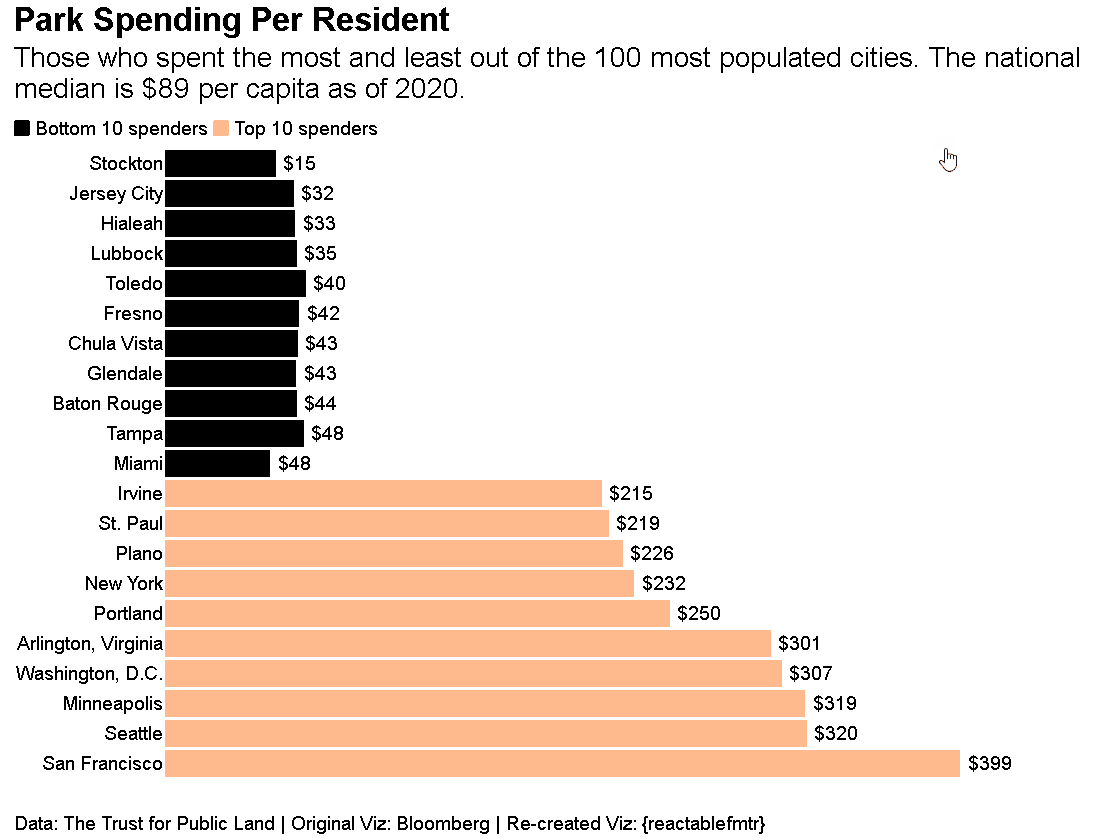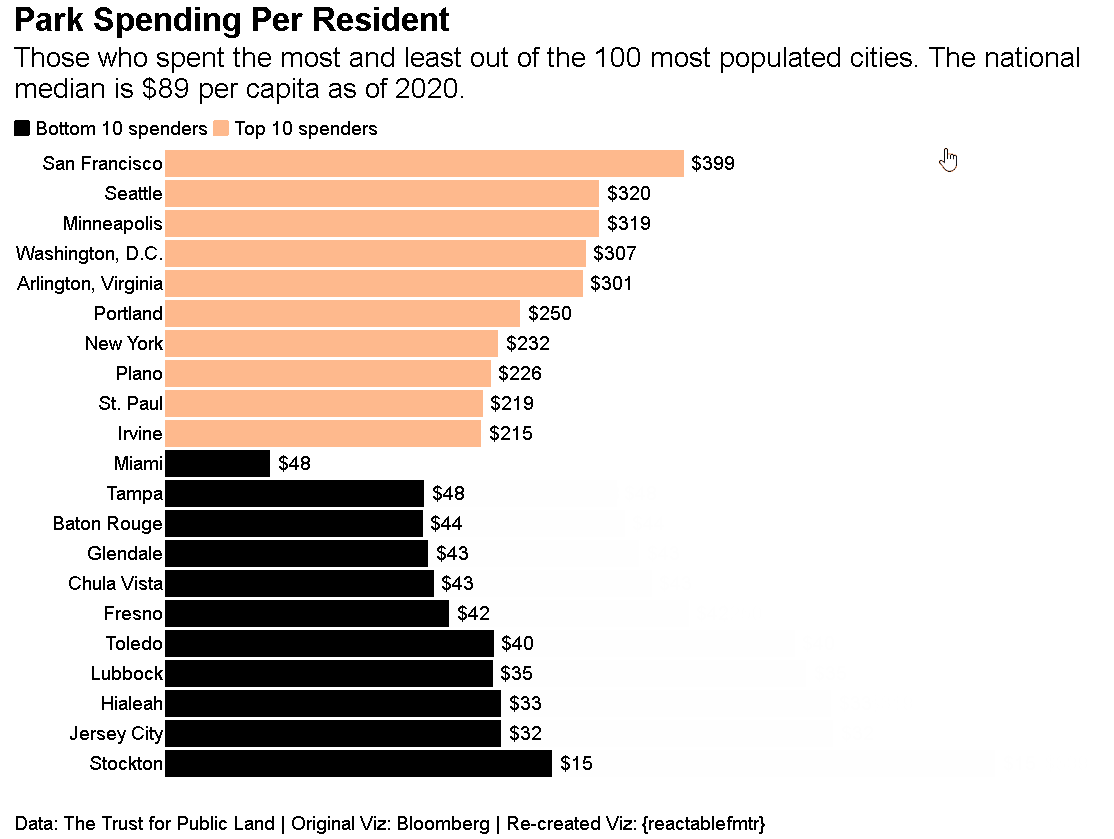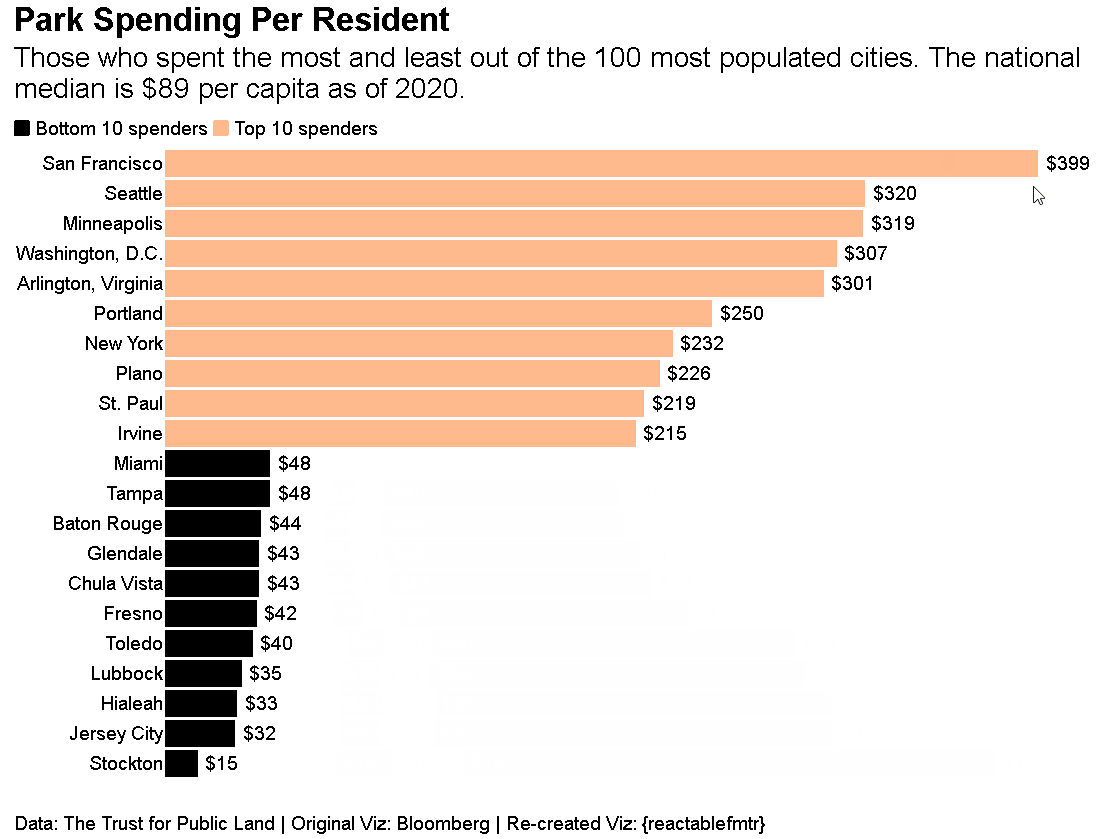
Who says {reactablefmtr} can just be used to make tables? You can create data visualizations like the one shown below. Source code
{reactablefmtr} or {reactable} tables can be saved directly to a file as a static PNG image or as an interactive HTML file with save_reactable().
Save as a PNG file:
save_reactable(table_name, "table.png")
Save as an HTML file:
save_reactable(table_name, "table.html")
If custom CSS styling is applied to the table within an R Markdown document:
save_reactable("table_name.Rmd", "table.png")
If you prefer to use a pipe:
table_name %>%
save_reactable("table.png")
A presentation given to the SoCal R Users Group: https://rpubs.com/kcuilla/reactablefmtr-socal-presentation
-
A huge thank you to Greg Lin for creating the amazing {reactable} package! Without Greg, {reactablefmtr} simply would not exist!
-
Thank you to Kent Russell for putting together the wonderful {dataui} package and suggesting integrating the code with {reactablefmtr} to allow interactive sparkline customization in reactable tables.
-
Last but not least, thank you to those below who have contributed to the development and/or documentation of the package! @yjunechoe, @shannonpileggi, @TimTeaFan