majianjia / nnom Goto Github PK
View Code? Open in Web Editor NEWA higher-level Neural Network library for microcontrollers.
License: Apache License 2.0
A higher-level Neural Network library for microcontrollers.
License: Apache License 2.0
Hi majianjia.
With your kind support of ZeroPadding layer, I deployed my caffe model successfully.
Now I am trying to deploy my model on memory constraint chip, and I found ZeroPadding layer consume much memory.
#3 Conv2D - ReLU - ( 64, 64, 8) 819k (17,424,32,768, 100) 1 1 1 - - - - -
#4 ZeroPad - - ( 65, 65, 8) (32,768,33,800, 0) 1 - 1 - - - - -
I think ZeroPadding and Cropping can be replaced as Single Buffer layer, not sure though.
I expect a quick response as before.
Thanks again for your great work.
Hi, this is a wonderful project.
Actually I just noticed that Tensorflow team just released a MCU version that can run TFLite model on microcontrollers.
https://www.sparkfun.com/products/15170
Hello, I was trying to use a model wide-resnet (an implementation of the model resnet_v1 in https://keras.io/zh/examples/cifar10_resnet/ with the parameter n=1 and num_filters = 8). I was using the version of nnom with cmsis-nn.
When i run the model (model_run(model);), it returns me an error NN_SIZE_MISMATCH.
Using the debugger, i have found the issue. In the function nnom_status_t conv2d_run(nnom_layer_t *layer) of the file nnom_conv2d.c, the function arm_convolve_1x1_HWC_q7_fast_nonsquare is used. This function returns NN_SIZE_MISMATCH if the strides parameters are not equal to 1 (also if the pad parameters are not equals to 0 or the kernel size is not equal to 1x1). CMSIS-NN doesn't support 1x1 convolution with strides!=1.
I suggest to modify the line of codes 293 of nnom_conv2d.c :
if (cl->kernel.w == 1 && cl->kernel.h == 1)
by:
if (cl->kernel.w == 1 && cl->kernel.h == 1 && cl->stride.w == 1 && cl->stride.h == 1 && cl->pad.w == 0 && cl->pad.h == 0)
in order to solve this issue.
thanks for your contributation,The question of me is: when the nets inference,why we all do not use batchnorm?whether your all nets-mcu project do not use batchnorm ?
I just do a project,my nets use batchnorm,but when I move to MCU,I find much Calculated amount added,what can I do?thank you
还有建议做成arduino库,用的人会超级多
Hello, great library you have here! I was looking for something exactly like this! I am currently using v0.2.0_beta.
I am trying to convert a Keras model to compile on ARM Cortex M7 with CMSIS NN optimization enabled, but I am running to this issue during compilation:
../source/weight.h:76:2: error: #error DENSE_2_OUTPUT_RSHIFT must be bigger than 0
Looking at the generated weight file, I find the lines:
#define DENSE_1_OUTPUT_SHIFT 0
// ......
#define DENSE_2_KERNEL_0_SHIFT (6)
// ......
#define DENSE_2_OUTPUT_SHIFT 7
// ......
#define DENSE_2_OUTPUT_RSHIFT (DENSE_1_OUTPUT_SHIFT+DENSE_2_KERNEL_0_SHIFT-DENSE_2_OUTPUT_SHIFT)Which would obviously result in -1...
Additionally, while running the utility script to convert my Keras model, I also ran into the following issue in line 582 of nnom_utils.py as it tried to generate the last Dense layer with softmax activation.
Lines 578 to 582 in 3f24572
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-6-f285063f1d0f> in <module>()
6 print(model.layers)
7
----> 8 generate_model(model, x, name='weights.h')
/home/kelvinchan/kick-sensor-cnn/nnom/scripts/nnom_utils.py in generate_model(model, x_test, name)
580 fp.write('\tlayer[%s] = model.hook(Output(shape(%s,1,1), nnom_output_data), layer[%s]);\n'%(id+1, layer.input.shape[1], id))
581 else:
--> 582 fp.write('\tlayer[%s] = model.hook(Output(shape%s, nnom_output_data), layer[%s]);\n'%(id+1, layer.shape[1:], id))
583 fp.write('\tmodel_compile(&model, layer[0], layer[%s]);\n'%(id+1))
584 if(ID>32):
AttributeError: 'Dense' object has no attribute 'shape'By changing layer.shape[1:] to layer.input.shape[1:], I got it to successfully generate weights.h, but I'm not sure if it is the reason why I encounter the first issue.
My Keras model summary:
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv1d_1 (Conv1D) (None, 998, 64) 448
_________________________________________________________________
conv1d_2 (Conv1D) (None, 996, 64) 12352
_________________________________________________________________
dropout_1 (Dropout) (None, 996, 64) 0
_________________________________________________________________
max_pooling1d_1 (MaxPooling1 (None, 498, 64) 0
_________________________________________________________________
flatten_1 (Flatten) (None, 31872) 0
_________________________________________________________________
dense_1 (Dense) (None, 100) 3187300
_________________________________________________________________
dense_2 (Dense) (None, 8) 808
=================================================================
Total params: 3,200,908
Trainable params: 0
Non-trainable params: 3,200,908
_________________________________________________________________
Hello,
I have quantized my own model and am running it on a nRF52840 (Cortex-M4F). I am surprised that enabling the CMSIS-NN backend results in a significant performance downgrade (~23%):
Local backend performance log
CMSIS backend performance log
Apart for the first layer (which is the only one showing a performance increase), all other convolution and dense layers seem to be using the optimized versions of the CMSIS functions (arm_convolve_HWC_q7_fast_nonsquare / arm_fully_connected_q7_opt). Would this performance downgrade be expected in my use case for some reason?
For information, I had to comment-out the call to "arm_convolve_HWC_q15_basic_nonsquare" in nnom_conv2d.c:394 as I cannot seem to find any compatible function in the latest CMSIS-NN release, and it would cause a compile-time error.
However, the predictions with the CMSIS backend seem to be different than the local backend, and much closer to the actual predictions of the raw Keras model:
Local backend predictions
CMSIS backend perdictions
Keras model predictions
The CMSIS backend gives the correct predictions for all test samples. The local backend gives tied predictions for 6/22 samples and wrong predictions for 3/22 samples. Is this prediction performance difference expected? Also, is there a user-friendly way to tweak the quantization to allow some chosen saturation or should I modify nnom.py?
Thank you in advance !
Now I know nnom is a way to do guess on mcu.
We should train our model on keras.Then use script to generate model c files to our project.
Finally our mcu run the model and guess the result.
It is not a fully edge AI way.Is there a plan to make mcu train and create its model self?
I am looking forward to you reply.
I have a siamese model, try to use generate_model(model, x_val, name=weights), but i don't know how to transform data to x_val? my model input is ([img1, img2], label), img1 and img2 shape is (1, 30, 30, 1), label is [1,0]
Model: "model_17"
input_24 (InputLayer) (None, 30, 30, 1) 0
conv2d_22 (Conv2D) (None, 30, 30, 32) 320
max_pooling2d_22 (MaxPooling (None, 15, 15, 32) 0
conv2d_23 (Conv2D) (None, 15, 15, 32) 9248
Total params: 9,568
Trainable params: 9,568
Non-trainable params: 0
Model: "model_18"
input_22 (InputLayer) (None, 30, 30, 1) 0
input_23 (InputLayer) (None, 30, 30, 1) 0
model_17 (Model) (None, 7, 7, 32) 9568 input_22[0][0]
input_23[0][0]
subtract_8 (Subtract) (None, 7, 7, 32) 0 model_17[1][0]
model_17[2][0]
conv2d_24 (Conv2D) (None, 7, 7, 32) 9248 subtract_8[0][0]
max_pooling2d_24 (MaxPooling2D) (None, 3, 3, 32) 0 conv2d_24[0][0]
flatten_6 (Flatten) (None, 288) 0 max_pooling2d_24[0][0]
dense_12 (Dense) (None, 64) 18496 flatten_6[0][0]
Total params: 37,377
Trainable params: 37,377
Non-trainable params: 0
cutoff '.txt' is the real name:
siamese-0.981.h5.txt
Hi,
It appears that when you try to create a weight file for depthwise seperable conv layers there are a few problems
1.) Missing biases and output shifts - these appear to be missing for all seperable conv layers
2.) Re-declaration during weight declaration - the depthwise weights are immediately redeclared with the pointwise weights.
3.) Model Declaration Wrong - instead of using DW_Conv2D, the generated model architecture calls Conv2D
Not sure if this is a bug or a missing feature (i.e. depthwise separable are still under development)
Hello,
I have been trying to run the included examples but have ran into issues:
mnist_simple.py ran to completion after I applied the cudNN fix and renamed 'acc' and 'val_acc' to 'accuracy' and 'val_accuracy' respectively. I attach the log and self-evaluation results which are as expected. However, when I try to run the generated quantized network in C, the results are heavily skewed towards one label. I attach the run log, and a simple C file with Makefile (meant to be placed and ran from the mnist-simple directory) to reproduce the issue:
mnist-simple.zip
auto_test.py crashes during execution, here is the (anonymized) execution log:
auto_test_log.txt
Thank you in advance
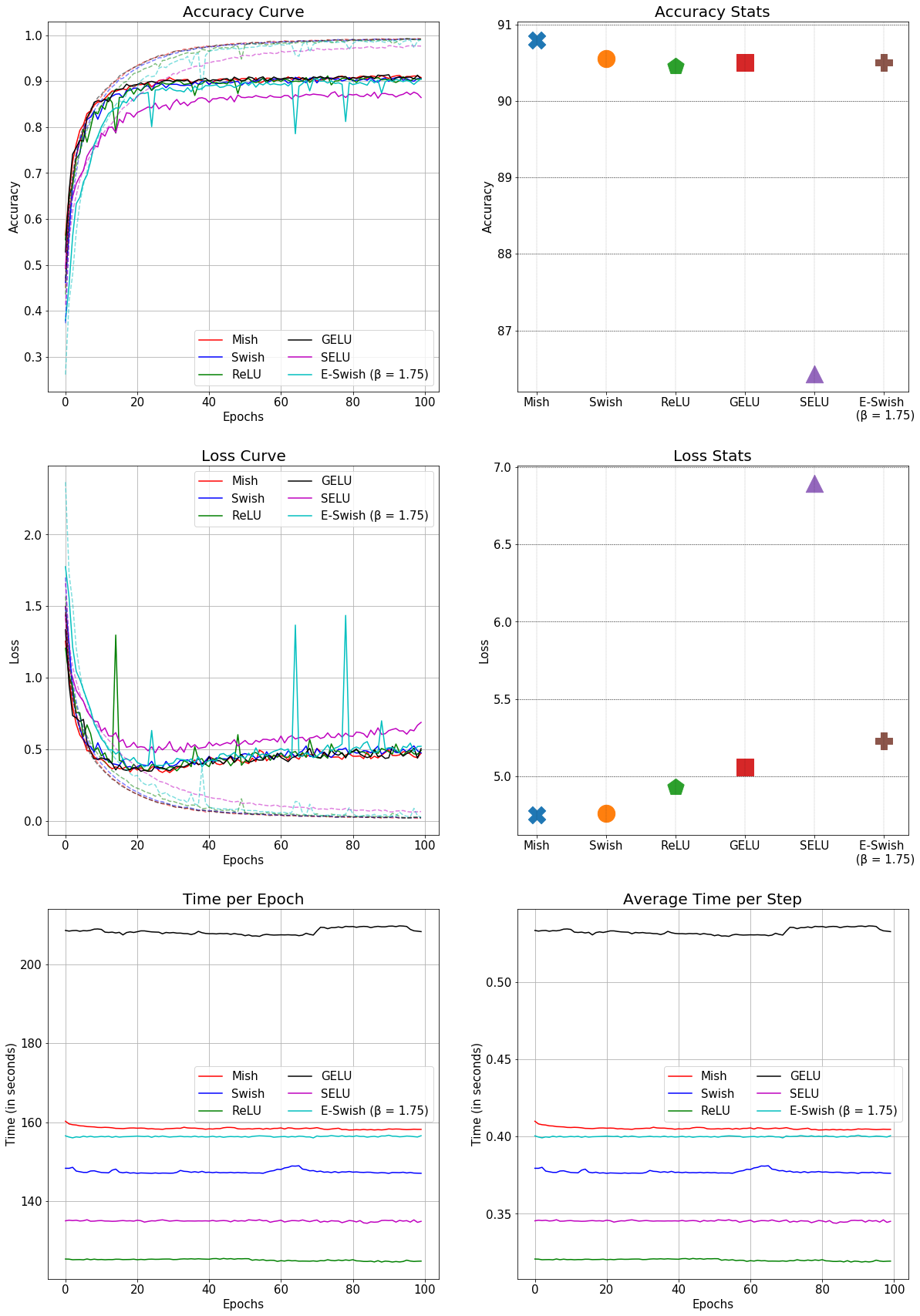
Mish is a new novel activation function proposed in this paper.
It has shown promising results so far and has been adopted in several packages including:
All benchmarks, analysis and links to official package implementations can be found in this repository
It would be nice to have Mish as an option within the activation function group.
This is the comparison of Mish with other conventional activation functions in a SEResNet-50 for CIFAR-10: (Better accuracy and faster than GELU)
我最近在做一个类似的嵌入式的项目,可以的话我想为nnom做一些开源的贡献。特别希望和您能有更深入的交流,有什么交流群之类的交流渠道吗?期待回复。
你好,请教下本工程可能转到安卓使用么
Hi,
I am seeing the difference in outputs of rnn-denoise for a noisy input file, when I compile with default provided "weights.h" file and "denoise_weights.h" file.
The Weights.h file has model VERSION number as below
/* model version /
#define NNOM_MODEL_VERSION (100000 + 100*4 + 2)
The denoise_weights.h file has model VERSION Number as below
/* model version /
#define NNOM_MODEL_VERSION (100000 + 100*4 + 3)
I see the performance of rnn-denoise output is degraded with V0.4.3(i.e. using denoise_weights.h header) compared to V0.4.2(i.e. weights.h header).
Could you please help me to find out why there is so much performance difference observed? Is there any difference in training data b/w 4.2 and 4.3?
Thanks,
Shiva
Hi Mr. Ma,
many thanks for your great work!
I evaluate nnom on windows for later use on MCUs, but I have a problem with your KWS example. All values of the dense output are max values (127 as int).
When I was looking for the cause, I wondered if the quantisation of 32bit audio to 16bit audio with
SaturaLH((p_raw_audio[i] >> 8)*1, -32768, 32767);should be
p_raw_audio[i] >> 16;instead?
Hi,
thanks for your great work!.
Do you have any ideas on why the prediction is always wrong when the NNOM_USING_CMSIS_NN is defined ?.
Without that optimization, the predictions are correct:
While in that case I always get the output predictions (4 classes with Softmax) always the same value: 32.
My model is a ResNet with 1D CNN and 1D max pool.
This is how I used the API: generate_model(model, x_test, name='quant_model.h', format='hwc').
我的设备为stm32h743vit6,核心板来自:https://gitee.com/WeAct-TC/MiniSTM32H7xx。
环境为cubemx生成hal库,然后导入mdk进行编译的。
然后运行报错告诉我网络结构中的Conv2d参数少了一个,我查看源代码后发现相较于教程中多了一个dilation参数,如图:
然后我查看了添加了keras文档后其默认的dilation参数为(1, 1),所以我直接复制了前面的stride(1, 1)作为其参数(虽然我不知道是否正确),如图:

之后便可以不报错烧录进单片机,但是我通过串口结果发现运行到Input层到p5点的时候就停止运行了:
p5点我设置在compile_layer函数的输出dim的地方:

我看代码里dilation也是nnom_3d_shape_t类的,不知道哪里出问题了。
Hi in the doc is write https://majianjia.github.io/nnom/api_layers/#output
nnom_layer_t* Output(nnom_3d_shape_t output_shape* p_buf);
but the type of Output is
nnom_layer_t *Output(nnom_3d_shape_t output_shape, void *p_buf)
Thanks you for your work!
Using the Keras to NNoM converter, I got following compiler error(s):
In file included from ../src/main.c:19:
../include/model.h:55:70: error: 'CONV2D_OUTPUT_RSHIFT' undeclared here (not in a function); did you mean 'CONV2D_OUTPUT_SHIFT'?
55 | static const nnom_weight_t conv2d_w = { (const void*)conv2d_weights, CONV2D_OUTPUT_RSHIFT};
| ^~~~~~~~~~~~~~~~~~~~
| CONV2D_OUTPUT_SHIFT
../include/model.h:57:90: error: 'DEPTHWISE_CONV2D_OUTPUT_RSHIFT' undeclared here (not in a function); did you mean 'DEPTHWISE_CONV2D_OUTPUT_SHIFT'?
57 | static const nnom_weight_t depthwise_conv2d_w = { (const void*)depthwise_conv2d_weights, DEPTHWISE_CONV2D_OUTPUT_RSHIFT};
| ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| DEPTHWISE_CONV2D_OUTPUT_SHIFT
../include/model.h:59:94: error: 'DEPTHWISE_CONV2D_1_OUTPUT_RSHIFT' undeclared here (not in a function); did you mean 'DEPTHWISE_CONV2D_1_OUTPUT_SHIFT'?
59 | static const nnom_weight_t depthwise_conv2d_1_w = { (const void*)depthwise_conv2d_1_weights, DEPTHWISE_CONV2D_1_OUTPUT_RSHIFT};
| ^~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| DEPTHWISE_CONV2D_1_OUTPUT_SHIFT
In file included from ../src/main.c:19:
../include/model.h: In function 'nnom_model_create':
../include/model.h:75:25: error: expected expression before ']' token
75 | layer[0] = Input(shape[], nnom_input_data);
| ^
../include/model.h:76:90: error: 'conv2d_b' undeclared (first use in this function); did you mean 'conv2d_w'?
76 | layer[1] = model.hook(Conv2D(8, kernel(1, 128), stride(1, 1), PADDING_SAME, &conv2d_w, &conv2d_b), layer[0]);
| ^~~~~~~~
| conv2d_w
../include/model.h:76:90: note: each undeclared identifier is reported only once for each function it appears in
../include/model.h:77:103: error: 'depthwise_conv2d_b' undeclared (first use in this function); did you mean 'depthwise_conv2d_w'?
77 | layer[2] = model.hook(DW_Conv2D(1, kernel(64, 1), stride(1, 1), PADDING_VALID, &depthwise_conv2d_w, &depthwise_conv2d_b), layer[1]);
| ^~~~~~~~~~~~~~~~~~
| depthwise_conv2d_w
../include/model.h:79:104: error: 'depthwise_conv2d_1_b' undeclared (first use in this function); did you mean 'depthwise_conv2d_1_w'?
79 | layer[5] = model.hook(DW_Conv2D(1, kernel(1, 16), stride(1, 1), PADDING_SAME, &depthwise_conv2d_1_w, &depthwise_conv2d_1_b), layer[4]);
| ^~~~~~~~~~~~~~~~~~~~
| depthwise_conv2d_1_w
The nnom_model_create function looks like this:
static nnom_model_t* nnom_model_create(void)
{
static nnom_model_t model;
nnom_layer_t* layer[11];
new_model(&model);
layer[0] = Input(shape[], nnom_input_data);
layer[1] = model.hook(Conv2D(8, kernel(1, 128), stride(1, 1), PADDING_SAME, &conv2d_w, &conv2d_b), layer[0]);
layer[2] = model.hook(DW_Conv2D(1, kernel(64, 1), stride(1, 1), PADDING_VALID, &depthwise_conv2d_w, &depthwise_conv2d_b), layer[1]);
layer[4] = model.hook(AvgPool(kernel(1, 8), stride(1, 8), PADDING_VALID), layer[3]);
layer[5] = model.hook(DW_Conv2D(1, kernel(1, 16), stride(1, 1), PADDING_SAME, &depthwise_conv2d_1_w, &depthwise_conv2d_1_b), layer[4]);
layer[7] = model.hook(AvgPool(kernel(1, 8), stride(1, 8), PADDING_VALID), layer[6]);
layer[8] = model.hook(Dense(4, &dense_w, &dense_b), layer[7]);
layer[9] = model.hook(Softmax(), layer[8]);
layer[10] = model.hook(Output(shape(4,1,1), nnom_output_data), layer[9]);
model_compile(&model, layer[0], layer[10]);
return &model;
}
Where does this error comes from? How can I avoid them? Is it because of the use_bias = False which I set in both Conv2D and DepthwiseConv2D layer?
Hello my model have 1 output with a value from 0.0 to 1.0
x=Dense(1)(x)
predictions=Activation('sigmoid')(x)
then when my float model ouput are 1 my optimized model output are label=1 prob=1.0 but when my float model ouput are 0.99 my optimized model output are label=0 prob=0.0.
I do somthing wrong or is a bug? how to have a outpour form 0 to 255 or 0 to 127?
If you can help my thanks you.
Hello again,
I have been trying to implement a small 1D CNN binary classifier with sigmoid activation at the end. When I copy my test inputs to nnom_input_data and run nnom_predict with the compiled model, prob always output to 1 for some reason, even though I verified the model prediction results with the same test data on Keras.
Here is the summary of the network from Keras:
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) (None, 1000, 2) 0
_________________________________________________________________
conv1d_1 (Conv1D) (None, 991, 4) 84
_________________________________________________________________
max_pooling1d_1 (MaxPooling1 (None, 495, 4) 0
_________________________________________________________________
re_lu_1 (ReLU) (None, 495, 4) 0
_________________________________________________________________
conv1d_2 (Conv1D) (None, 489, 8) 232
_________________________________________________________________
max_pooling1d_2 (MaxPooling1 (None, 244, 8) 0
_________________________________________________________________
re_lu_2 (ReLU) (None, 244, 8) 0
_________________________________________________________________
conv1d_3 (Conv1D) (None, 240, 16) 656
_________________________________________________________________
max_pooling1d_3 (MaxPooling1 (None, 120, 16) 0
_________________________________________________________________
re_lu_3 (ReLU) (None, 120, 16) 0
_________________________________________________________________
global_max_pooling1d_1 (Glob (None, 16) 0
_________________________________________________________________
dense_1 (Dense) (None, 50) 850
_________________________________________________________________
re_lu_4 (ReLU) (None, 50) 0
_________________________________________________________________
dense_2 (Dense) (None, 1) 51
_________________________________________________________________
activation_1 (Activation) (None, 1) 0
=================================================================
Total params: 1,873
Trainable params: 1,873
Non-trainable params: 0
I am following the procedure from the mnist-simple example:
int8_t data[1000][2000];
uint32_t predict_label;
float prob = 0;
// load data from binary file
model = nnom_model_create();
memcpy(nnom_input_data, data[i], sizeof(nnom_input_data));
nnom_predict(model, &predict_label, &prob);
printf("Probability: %.2f \r\n", prob);Currently using v0.2.0 in the MCU code.
If anyone has any insights or advice, that'll be much appreciated.
To use RNN layers, must define NNOM_TUNCATE in nnom_port.h to use 'floor' instead of 'round' in backend ops.
Problem seems related to NNOM_ROUND() in
local_mult_q15() and local_fully_connected_mat_q7_vec_q15()
Same as ARM versions, NN_ROUND(). I need to review those shift and round calculation. At the meantime, please define NNOM_TUNCATE in nnom_port.h
Hi. I'm currently working around with tricore platform and trying to implement nnom on tc26x/tc37x. When porting I encountered forward declaration errors with typedef in nnom.h.
For example, in line 256 and 275 it used forward declaration like this:
// nn wrappers
typedef struct _nnom_mem_block_t nnom_mem_block_t;
typedef struct _nnom_buf
{
nnom_mem_block_t *mem;
size_t size;
uint8_t type;
} nnom_buf_t;
// a memory block to store pre-assign memories during compiling. then assigned to each tensor after.
typedef struct _nnom_mem_block_t
{
void *blk; // data block location
size_t size; // the maximum size for this block
uint8_t owners; // how many layers own this block
uint8_t state; // empty? filled? for static nn, currently only used in compiling
} nnom_mem_block_t;which is totally fine when i use nnom in stm32 projects. But when i used tricore's ctc compiler, it gave me errors like previous declaration of "nnom_mem_block_t".
So I digged it up a little and found a possible explanation is that the current style of forward declaration only works under C11 standard. Since infineon's ctc compiler only supports C99 in their free Aurix Development Studio, problem occurs.
After that I experimented on NXP IDE and STM32 IDE with gcc(c11) together with ADS and found some alternative style as follows.
//No.1 Valid usage for both gcc(c11) and tricore ctc(c99) compiler
typedef struct _student student_t;
struct _intern_a{
struct _student *stu;
};
struct _intern_b{
student_t *stu;
};
struct _student{
student_t *p;
int id;
};//No.2 Valid usage for both gcc(c11) and tricore ctc(c99) compiler
struct _student;
typedef struct _student{
struct _student *p;
int id;
}student_t;(Negative example: this won't work under C99 standard.)
//No.3 Invalid usage for tricore ctc(c99) compiler,but work just fine with gcc(c11) compiler
typedef struct _student student_t;
typedef struct _student{
student_t *p;
int id;
}student_t;I‘ve used first method to refactor nnom src, and now it seems to be just fine.
I think the current style is totally fine because mainstream chip manufacturers use C11 as their gcc's default C standard (In fact infineon provide TASKING toolsets who claims to support C11 standard, but it is too expensive for individuals), but considering maximum portability, is it necessary to refactor this part to adapt C99 standard?
Currently, the Q format is handled by the scripts.
The nnom cannot really know what is the q format of the layers outputs but their output shifting.
This is fine with most of the layers, such as conv, dense, and relu.
However, the activations such as sigmoid and tanh must know the current Q format because they are doing arithmetic base on the real number. There is a "num int bit" argument, which should be the m in Qmn format.
I am still thinking of the solution.
@parai
Do you have any suggestion?
Thanks
I think #include "layers/nnom_avgpool.h" is missing in nnom/inc/nnom_layers.h
If i add it it is ok
Hi @majianjia 👍
I try to build a simple neural network to predict the outputs of XOR logic like this:
And generate the "weights.h" use the following code:
finally, run the neural network:
#include "weights.h"
nnom_model_t* model;
void main(void)
{
model = nnom_model_create();
nnom_input_data[0] = 0;
nnom_input_data[1] = 0;
model_run(model);
nnom_input_data[0] = 0;
nnom_input_data[1] = 1;
model_run(model);
nnom_input_data[0] = 1;
nnom_input_data[1] = 0;
model_run(model);
nnom_input_data[0] = 1;
nnom_input_data[1] = 1;
model_run(model);
}The inputs are [0, 1] and the outputs should be [1] but it's [106]. so where I am wrong?
Hi jianjia,
I see the snip code in file (https://github.com/majianjia/nnom/blob/master/examples/keyword_spotting/kws.py) as below:
` def normalize(data, n, quantize=True):
limit = pow(2, n)
data = np.clip(data, -limit, limit) / limit
if quantize:
data = np.round(data * 128) / 128.0
return data
# instead of using maximum value for quantised, we allows some saturation to save more details in small values.
x_train = normalize(x_train, 3)
x_test = normalize(x_test, 3)
x_val = normalize(x_val, 3)`
You select [-8,8] as the range for the dataset.
I wonder how to decide the feature range for the dataset.
Thanks.
Hi @majianjia 👍
Here is what I though that to make nnom much more better and suitable for MCU.
I prefer to generated the model to a static const struct array which could be used to represent the model, for example as below demo code shows:
typedef enum {
L_CONV2D = 0,
L_RELU,
L_MAXPOOL,
....
} layer_type_t;
type struct {
layer_type_t type;
q7*_t weight_array[]; // weights and bias
q7* input; // input buffer: TODO multi-inputs
q7* output; // output buffer
shape_t shape... etc..
} layer_t;
q7_t buffer1[1024];
q7_t buffer2[1024];
static const layer_t model [] = {
{L_CONV2D, weghts, buffer1, buffer2, ... },
{L_RELU, weghts, buffer2, buffer1, ... },
....
};
int main(void) {
// loop and run each layer
for layer in model {
run this layer
}
}Hi @majianjia . Thank you fir the project you have developed, It is very handy.
I am trying to use the code developed here with a LSTM network.
I saw in
Line 38 in fc05b4a
nnom/examples/auto_test/main.py
Lines 121 to 127 in fc05b4a
Can you provide an example how to feed the input. I am giving the input to the generate_model similar to the way I am giving to predict() (or https://keras.io/api/models/model/) function of but it is throwing errors. I saw, in the example for the time series data , at the end rnn models are not used.
Thank you.
I met with a Error when running rnn-denoise example, "ValueError: Input 0 is incompatible with layer gru: expected shape=(2048, None, 40), found shape=[32, 1, 40]"
This Error occurs in the function voice_denoise: prediction = model.predict(feat)
And it also happens when generating weight file (model.predict also used there).
After I checked tensorflow/python/keras/engine/training.py, I modified main.py from "prediction = model.predict(feat)" to "prediction = model.predict(feat,batch_size=timestamp_size)". It seemed OK now.
So, is this modification OK or not? And if it's OK, then how to modify nnom.py?
Hi,
I try to train the rnn-denoise demo, but it failed when generating the NNoM model in quantize_output function. Is there any update for this demo?
Here is the log:
filtering with frequency: 5394.2623288811965
filtering with frequency: 6162.04951128775
filtering with frequency: 7026.566556252747
/root/miniconda3/envs/myconda/lib/python3.5/site-packages/matplotlib/figure.py:448: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
% get_backend())
input_1 Quantized method: max-min Values max: 1.0 min: -1.0 dec bit 7
================
[<tensorflow.python.keras.engine.input_layer.InputLayer object at 0x7fd630553ac8>]
Traceback (most recent call last):
File "main.py", line 315, in <module>
main()
File "main.py", line 297, in main
generate_model(model, x_train[:timestamp_size*4], name='denoise_weights.h')
File "/mnt/nnom/scripts/nnom.py", line 750, in generate_model
layer_q_list = quantize_output(model, x_test, layer_offset=False, quantize_method=quantize_method)
File "/mnt/nnom/scripts/nnom.py", line 498, in quantize_output
layer_model = Model(inputs=model.input, outputs=in_layer.output)
AttributeError: 'list' object has no attribute 'output'
(myconda) root@f8209e3ef1a3:/mnt/nnom/examples/rnn-denoise#
B&R
I give the same input(512 zero samples.) to the 'mfcc‘ api in main.c and python with the same setting, but i got different results. The setting is shown below:
Python: 
mfcc api in main.c  
The input is 512 zero sample, and the result is -84.3408 0.0000 -0.0000 0.0000 -0.0000 0.0000 -0.0000 0.0000 -0.0000 -0.0000 0.0001 -0.0001 0.0000 -0.0000 -0.0001 -0.0000 0.0000 0.0001 -0.0001 0.0002 in main.c and '-36.0437,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000,0.0000' in python.
The mfcc setting is the same as original rnn-denoise example that makes me confused a lot.
I will appreciate a lot for any help.
Traceback (most recent call last):
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\runpy.py", line 87, in _run_code
exec(code, run_globals)
File "c:\Users\auma-\.vscode\extensions\ms-python.python-2021.4.765268190\pythonFiles\lib\python\debugpy\__main__.py", line 45, in <module>
cli.main()
File "c:\Users\auma-\.vscode\extensions\ms-python.python-2021.4.765268190\pythonFiles\lib\python\debugpy/..\debugpy\server\cli.py", line 444, in main
run()
File "c:\Users\auma-\.vscode\extensions\ms-python.python-2021.4.765268190\pythonFiles\lib\python\debugpy/..\debugpy\server\cli.py", line 285, in run_file
runpy.run_path(target_as_str, run_name=compat.force_str("__main__"))
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\runpy.py", line 265, in run_path
return _run_module_code(code, init_globals, run_name,
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\runpy.py", line 97, in _run_module_code
_run_code(code, mod_globals, init_globals,
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\runpy.py", line 87, in _run_code
exec(code, run_globals)
File "c:\VisualRepos\nnom-master\examples\rnn-denoise\main.py", line 306, in <module>
main()
File "c:\VisualRepos\nnom-master\examples\rnn-denoise\main.py", line 292, in main
filtered_sig = voice_denoise(sig, rate, model, timestamp_size, numcep=y_train.shape[-1], plot=True) # use plot=True argument to see the gains/vad
File "c:\VisualRepos\nnom-master\examples\rnn-denoise\main.py", line 84, in voice_denoise
prediction = model.predict(feat, batch_size=timestamp_size)
File "C:\Users\auma-\anaconda3\envs\noise_reduce\lib\site-packages\tensorflow\python\keras\engine\training.py", line 1644, in predict
raise ValueError('Expect x to be a non-empty array or dataset.')
ValueError: Expect x to be a non-empty array or dataset.
I followed all of the previous information.
But when I run main.py, I get an error. What is the problem?
Please help me
Hi @majianjia 👍
When I try a simple code like than:
import numpy as np
import keras
import nnom_utils
x_train = np.array([[0, 0],
[0, 1],
[1, 0],
[1, 1]])
y_label = np.array([0, 1, 1, 0])
model = keras.Sequential()
model.add(keras.layers.Dense(8, input_shape=(2,), activation='relu'))
model.add(keras.layers.Dense(8, activation='relu'))
model.add(keras.layers.Dense(1, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy')
model.fit(x_train, y_label, batch_size=1, epochs=100)
nnom_utils.generate_model(model, x_train * 0.9)I got an error:
AttributeError: 'Dense' object has no attribute 'shape'
Hello,
For the generate_test_bin() utility function, the documentation says to provide a quantized input x into the utility function, but when I look at the script, it seems to already quantize the input.
Lines 67 to 80 in 3f24572
My current x_test are already floating values between (0~1). Do I still need to multiply by 127 (2*7 - 1) to first convert to Q7 before using generate_test_bin(), or should I leave it as is?
我使用了CMSIS-NN,在 port.h中也开启了,并且版本都是符合的,但是在实验中,使用了NN却没有提升太多的速度,比如不使用预测需要230 tick,使用后变为190 tick, 而在我自己写的神经网络中,不使用NN预测需要70 tick,使用后变为51 tick。 使用的硬件为stm32f407zet6,环境都是搭好的,rt-thread中1s 1000tick
Wonderful project!
I have one issue.
When I generate model, I got following error.
Traceback (most recent call last):
File "keras_to_nnom.py", line 18, in
generate_model(model, np.ones(shape=(1, 128, 128, 1)), name=weights)
File "/mnt/d/NUC970/Proj/nnom-master/scripts/nnom_utils.py", line 546, in generate_model
inps = [input.name.replace(':','/').split('/')[0] for input in layer.input]
File "/mnt/d/NUC970/Proj/nnom-master/scripts/nnom_utils.py", line 546, in
inps = [input.name.replace(':','/').split('/')[0] for input in layer.input]
File "/home/turing/.local/lib/python3.6/site-packages/tensorflow/python/framework/ops.py", line 442, in iter
"Tensor objects are only iterable when eager execution is "
TypeError: Tensor objects are only iterable when eager execution is enabled. To iterate over this tensor use tf.map_fn.
I am using tensorflow 1.13.1
Hope to any suggentions.
Hi Majianjia ,
I tested NNOM and ran into such a problem. I created a primitive model for the implementation of the XOR problem.

After train and generate model the values appear to be OK


After test on STM32G431 I found this problem (input is one dim vector as created in jupyter => input shape(2,) )

and output from console is

I tried to modify the code to test the functionality

after run the code output is OK ane value is ok too.

My question is , can work NNOM with input shape=(2,) ?
Thank you , nnom is excellent job :)
Hi @majianjia.
Thank you for your quick response everytime.
I have had accuracy test of my model using your framework.
It had got 99.2% using by caffe framework, but in nnom, it dropped to 95%.
Is there any way to improve this?
I've used ncnn int8 many times, and it's accuracy performance was quite good.
What do you think about their quantization method?
https://github.com/BUG1989/caffe-int8-convert-tools
I want your opinion about this problem.
Thanks.
Hello,
I am running into an error when running the quantization script on a model generated with tensorflow 2.3.1:
quantisation list {'input_1': [0, 0], 'conv1d': [1, 0], 'conv1d_1': [0, 0], 'conv1d_2': [1, 0], 'max_pooling1d': [1, 0], 'conv1d_3': [2, 0], 'conv1d_4': [2, 0], 'conv1d_5': [3, 0], 'max_pooling1d_1': [3, 0], 'flatten': [3, 0], 'dropout': [3, 0], 'dense': [2, 0], 'dropout_1': [2, 0], 'dense_1': [1, 0], 'dense_2': [0, 0]} quantizing weights for layer conv1d tensor_conv1d_kernel_0 dec bit 8 Traceback (most recent call last): File "main_keras.py", line 584, in <module> main() File "main_keras.py", line 581, in main port_to_uc() File "main_keras.py", line 575, in port_to_uc nnom.generate_model(classifier.model, classifier.validate_set.get_all_inputs(), name='weights.h') File "D:\Projects\FNH\deep_learning\nnom\scripts\nnom.py", line 746, in generate_model quantize_weights(model, per_channel_quant=per_channel_quant, name=name, format=format, layer_q_list=layer_q_list) File "D:\Projects\FNH\deep_learning\nnom\scripts\nnom.py", line 631, in quantize_weights layer_input_dec = layer_q_list[inp][0] KeyError: 'input_1_1'
It seems the input of the first actual layer (conv1d) is named "input_1_1", not "input_1" as in the quantisation list. Hacking the script to substitute the correct string allows it to keep running (until it hits a similar error on line 940).
I have tried a lot of time for using bias on Conv2d.(take weights at ckpt trained with tensorflow_1.15)
(very dirty code....sorry)

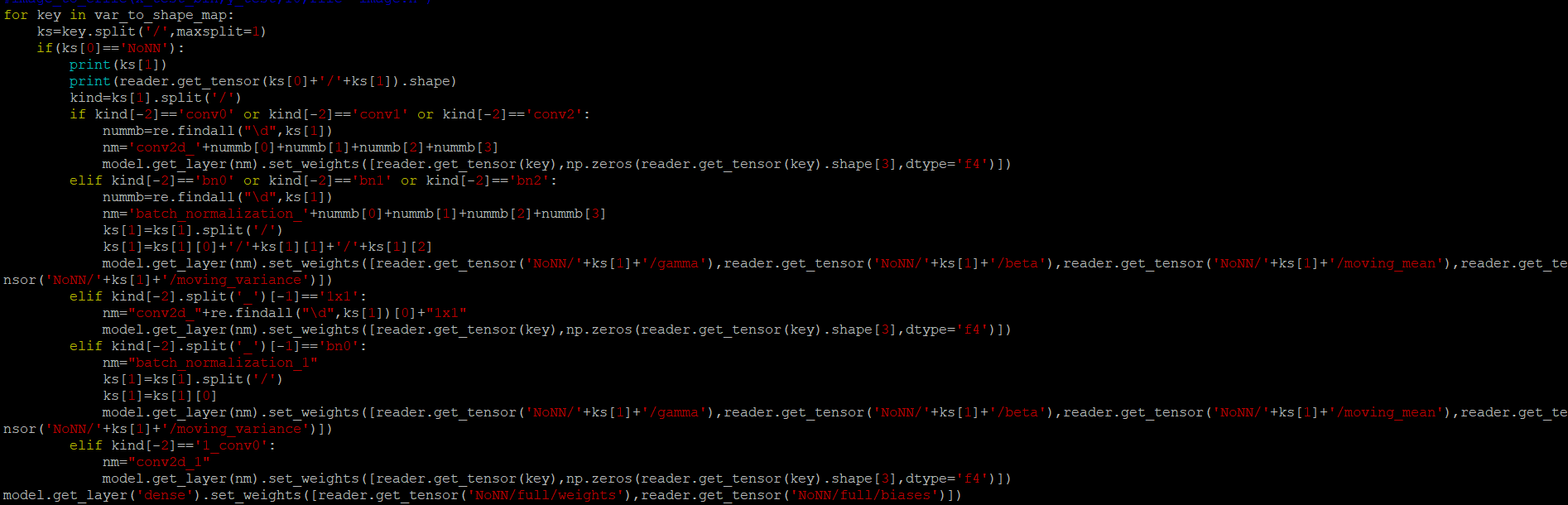
First I changed nnom_utils.py. Erased all c_b line, and Changed layer.set_weights([c_w]).
And Changed related Length of convolution layer.
Twice I added zeros bias, But np.log2 functions make error.
So I added if statement like 'if min_value'==0: min_value=1e-36'
Both ways make this problem.
The First evaluate_model function works fine
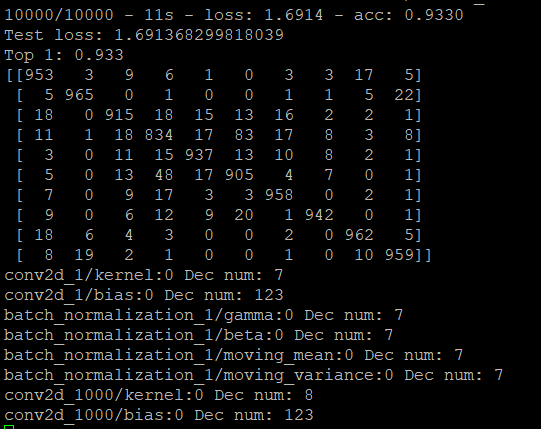
But if I do the generate_model function and then do the evaluate function again, the evaluate_model function doesn't work fine.
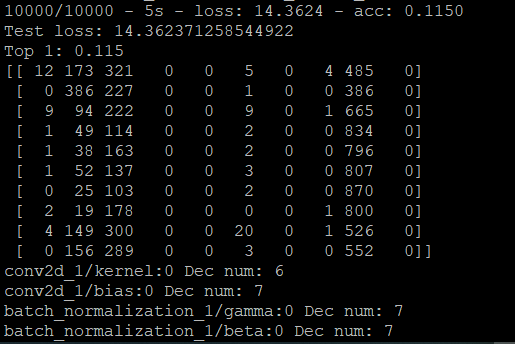
And I'll show you a picture of the results on the board.
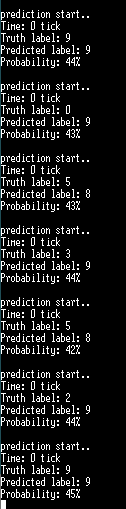
Thank you for reading the long article, and I apologize for terrible English and coding skills.
Safe Safety.
Running into the following error while trying to use nnom.generate_model(model, test_data)
Traceback (most recent call last):
File "C:/Users/Technerder/Dev/TimeSeriesClassification/train_lstm_1d.py", line 60, in <module>
nnom.generate_model(model, test_data)
File "C:\Users\Technerder\Dev\TimeSeriesClassification\nnom.py", line 746, in generate_model
layer_q_list = quantize_output(model, x_test, layer_offset=False, quantize_method=quantize_method)
File "C:\Users\Technerder\Dev\TimeSeriesClassification\nnom.py", line 527, in quantize_output
dec_bits = find_dec_bits_max_min(features, bit_width=8)
File "C:\Users\Technerder\Dev\TimeSeriesClassification\nnom.py", line 214, in find_dec_bits_max_min
max_val = abs(data.max()) - abs(data.max()/pow(2, bit_width)) # allow very small saturation.
AttributeError: 'BatchDataset' object has no attribute 'max'
Does nnom not support the usage of batched datasets?
I managed to get the keyword_spotting example running on ESP32 (ESP32-PICO-D4 of M5StickC), as a MicroPython module. It works great, except that sometimes it crashes in tensor_size(). Here is the call stack:
0x401f0612: tensor_size at ??:?
0x40118b29: input_run at ??:?
0x40118a69: model_run_to at ??:?
0x40118a89: model_run at ??:?
0x40117cc4: nnom_predict at ??:?
Note that the file names and line numbers are not available, as the sources are compiled using -Ofast to maximize speed optimization.
Seems there's a bug somewhere in the code. Do we have idea what that could be?
For a project, i have trained 2 CNN (Lenet and resnet) on the dataset GTSRB. Then I used Nnom to put them on microcontrollers.
The accuracy of Lenet is 99,4% on tensorflow and 99,1% on microcontroller.
But the accuracy of resnet is 99.3% on tensorflow and around 50% on microcontroller.
Can you look into my project (https://github.com/BaptisteNguyen/testNNom) in order to explain this difference?
I have test the demo on ubuntu16.04, but I got a bad result, some voice was cutoff and the noise of keyboard was not removed. while test with rnnoise, it got a good result.
I just checkout the code and run scons to make the demo, the input file was 16k 16bit mono noisy voice file.
The question is :
WARNING: model returned at #0
I followed your example auto_test with my own depthwise deparable CNN. After a few epochs of training my Keras model has an accuracy of 98.12% on the MNIST test set. After quantization the NNoM model has an accuracy of 12.95%. I do expect some performance drop but this is such a large drop that I rather think it is a bug. Here is the model summary:
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) (None, 28, 28, 1) 0
_________________________________________________________________
conv2d_1 (Conv2D) (None, 26, 26, 12) 120
_________________________________________________________________
batch_normalization_1 (Batch (None, 26, 26, 12) 48
_________________________________________________________________
activation_1 (Activation) (None, 26, 26, 12) 0
_________________________________________________________________
depthwise_conv2d_1 (Depthwis (None, 24, 24, 12) 120
_________________________________________________________________
batch_normalization_2 (Batch (None, 24, 24, 12) 48
_________________________________________________________________
activation_2 (Activation) (None, 24, 24, 12) 0
_________________________________________________________________
conv2d_2 (Conv2D) (None, 24, 24, 16) 208
_________________________________________________________________
batch_normalization_3 (Batch (None, 24, 24, 16) 64
_________________________________________________________________
activation_3 (Activation) (None, 24, 24, 16) 0
_________________________________________________________________
max_pooling2d_1 (MaxPooling2 (None, 12, 12, 16) 0
_________________________________________________________________
depthwise_conv2d_2 (Depthwis (None, 10, 10, 16) 160
_________________________________________________________________
batch_normalization_4 (Batch (None, 10, 10, 16) 64
_________________________________________________________________
activation_4 (Activation) (None, 10, 10, 16) 0
_________________________________________________________________
conv2d_3 (Conv2D) (None, 10, 10, 32) 544
_________________________________________________________________
batch_normalization_5 (Batch (None, 10, 10, 32) 128
_________________________________________________________________
activation_5 (Activation) (None, 10, 10, 32) 0
_________________________________________________________________
depthwise_conv2d_3 (Depthwis (None, 8, 8, 32) 320
_________________________________________________________________
batch_normalization_6 (Batch (None, 8, 8, 32) 128
_________________________________________________________________
activation_6 (Activation) (None, 8, 8, 32) 0
_________________________________________________________________
conv2d_4 (Conv2D) (None, 8, 8, 64) 2112
_________________________________________________________________
batch_normalization_7 (Batch (None, 8, 8, 64) 256
_________________________________________________________________
activation_7 (Activation) (None, 8, 8, 64) 0
_________________________________________________________________
depthwise_conv2d_4 (Depthwis (None, 6, 6, 64) 640
_________________________________________________________________
batch_normalization_8 (Batch (None, 6, 6, 64) 256
_________________________________________________________________
activation_8 (Activation) (None, 6, 6, 64) 0
_________________________________________________________________
conv2d_5 (Conv2D) (None, 6, 6, 96) 6240
_________________________________________________________________
batch_normalization_9 (Batch (None, 6, 6, 96) 384
_________________________________________________________________
activation_9 (Activation) (None, 6, 6, 96) 0
_________________________________________________________________
global_max_pooling2d_1 (Glob (None, 96) 0
_________________________________________________________________
dropout_1 (Dropout) (None, 96) 0
_________________________________________________________________
dense_1 (Dense) (None, 96) 9312
_________________________________________________________________
re_lu_1 (ReLU) (None, 96) 0
_________________________________________________________________
dropout_2 (Dropout) (None, 96) 0
_________________________________________________________________
dense_2 (Dense) (None, 10) 970
_________________________________________________________________
activation_10 (Activation) (None, 10) 0
=================================================================
Total params: 22,122
Trainable params: 21,434
Non-trainable params: 688
When I used nnom to transform my model into c source file, it failed to generate the model because of KeyError (could not find correct layer name).
I've checked my source and found that nnom is ok only when there is one model.fit(). If I run more than one fit() on one model, it crashed.
So I've checked the keras layer naming mechanism. It appears that keras names the layer automatically with the pattern: "the-layer-type_the-order-of-this-type-of-layer_fit-times-if-you-run-fit()-more-than-once)" ,but currently nnom only recongnize the first two part and has no regex to match "_fit-time".
For example, the name of the layer is actually "input_1_2", but nnom only get "input_1"
I think it can be solved by changing the name acquiring method of LI[].
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.