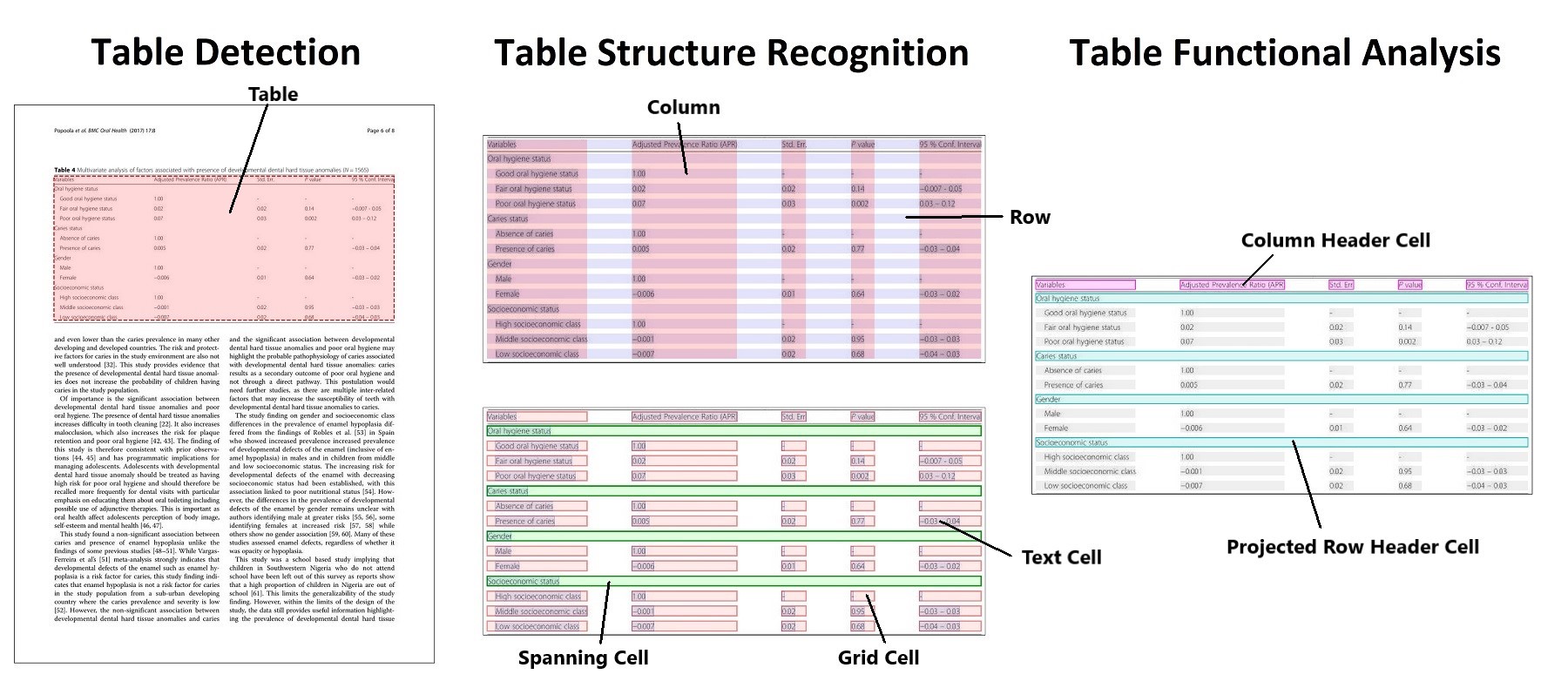
A deep learning model based on object detection for extracting tables from PDFs and images.
First proposed in "PubTables-1M: Towards comprehensive table extraction from unstructured documents".
This repository also contains the official code for these papers:
- "GriTS: Grid table similarity metric for table structure recognition"
- "Aligning benchmark datasets for table structure recognition"
Note: If you are looking to use Table Transformer to extract your own tables, here are some helpful things to know:
- TATR can be trained to work well across many document domains and everything needed to train your own model is included here. But at the moment pre-trained model weights are only available for TATR trained on the PubTables-1M dataset. (See the additional documentation for how to train your own multi-domain model.)
- TATR is an object detection model that recognizes tables from image input. The inference code built on TATR needs text extraction (from OCR or directly from PDF) as a separate input in order to include text in its HTML or CSV output.
Additional information about this project for both users and researchers, including data, training, evaluation, and inference code is provided below.
08/22/2023: We have released 3 new pre-trained models for TATR-v1.1 (trained on 1. PubTables-1M, 2. FinTabNet.c, and 3. both datasets combined) according to the details in our paper.
04/19/2023: Our latest papers (link and link) have been accepted at ICDAR 2023.
03/09/2023: We have added more image cropping to the official training script (like we do in our most recent paper) and updated the code and environment.yml to use Python 3.10.9, PyTorch 1.13.1, and Torchvision 0.14.1, among others.
03/07/2023: We have released a new simple inference pipeline for TATR. Now you can easily detect and recognize tables from images and convert them to HTML or CSV.
03/07/2023: We have released a collection of scripts to create training data for TATR and to canonicalize pre-existing datasets, such as FinTabNet and SciTSR.
03/01/2023: New paper "Aligning benchmark datasets for table structure recognition" is now available on arXiv.
11/25/2022: We have made the full PubTables-1M dataset alternatively available for download from Hugging Face.
05/05/2022: We have released the pre-trained weights for the table structure recognition model trained on PubTables-1M.
03/23/2022: Our paper "GriTS: Grid table similarity metric for table structure recognition" is now available on arXiv
03/04/2022: We have released the pre-trained weights for the table detection model trained on PubTables-1M.
03/03/2022: "PubTables-1M: Towards comprehensive table extraction from unstructured documents" has been accepted at CVPR 2022.
11/21/2021: Our updated paper "PubTables-1M: Towards comprehensive table extraction from unstructured documents" is available on arXiv.
10/21/2021: The full PubTables-1M dataset has been officially released on Microsoft Research Open Data.
06/08/2021: Initial version of the Table Transformer (TATR) project is released.
The goal of PubTables-1M is to create a large, detailed, high-quality dataset for training and evaluating a wide variety of models for the tasks of table detection, table structure recognition, and functional analysis.
It contains:
- 575,305 annotated document pages containing tables for table detection.
- 947,642 fully annotated tables including text content and complete location (bounding box) information for table structure recognition and functional analysis.
- Full bounding boxes in both image and PDF coordinates for all table rows, columns, and cells (including blank cells), as well as other annotated structures such as column headers and projected row headers.
- Rendered images of all tables and pages.
- Bounding boxes and text for all words appearing in each table and page image.
- Additional cell properties not used in the current model training.
Additionally, cells in the headers are canonicalized and we implement multiple quality control steps to ensure the annotations are as free of noise as possible. For more details, please see our paper.
We provide different pre-trained models for table detection and table structure recognition.
Table Detection:
| Model | Training Data | Model Card | File | Size |
|---|---|---|---|---|
| DETR R18 | PubTables-1M | Model Card | Weights | 110 MB |
Table Structure Recognition:
| Model | Training Data | Model Card | File | Size |
|---|---|---|---|---|
| TATR-v1.0 | PubTables-1M | Model Card | Weights | 110 MB |
| TATR-v1.1-Pub | PubTables-1M | Model Card | Weights | 110 MB |
| TATR-v1.1-Fin | FinTabNet.c | Model Card | Weights | 110 MB |
| TATR-v1.1-All | PubTables-1M + FinTabNet.c | Model Card | Weights | 110 MB |
Table Detection:
| Model | Test Data | AP50 | AP75 | AP | AR |
|---|---|---|---|---|---|
| DETR R18 | PubTables-1M | 0.995 | 0.989 | 0.970 | 0.985 |
Table Structure Recognition:
| Model | Test Data | AP50 | AP75 | AP | AR | GriTSTop | GriTSCon | GriTSLoc | AccCon |
|---|---|---|---|---|---|---|---|---|---|
| TATR-v1.0 | PubTables-1M | 0.970 | 0.941 | 0.902 | 0.935 | 0.9849 | 0.9850 | 0.9786 | 0.8243 |
PubTables-1M is available for download from Microsoft Research Open Data.
We have also uploaded the full set of archives to Hugging Face.
The dataset on Microsoft Research Open Data comes in 5 tar.gz files:
- PubTables-1M-Image_Page_Detection_PASCAL_VOC.tar.gz: Training and evaluation data for the detection model
/images: 575,305 JPG files; one file for each page image/train: 460,589 XML files containing bounding boxes in PASCAL VOC format/test: 57,125 XML files containing bounding boxes in PASCAL VOC format/val: 57,591 XML files containing bounding boxes in PASCAL VOC format
- PubTables-1M-Image_Page_Words_JSON.tar.gz: Bounding boxes and text content for all of the words in each page image
- One JSON file per page image (plus some extra unused files)
- PubTables-1M-Image_Table_Structure_PASCAL_VOC.tar.gz: Training and evaluation data for the structure (and functional analysis) model
/images: 947,642 JPG files; one file for each page image/train: 758,849 XML files containing bounding boxes in PASCAL VOC format/test: 93,834 XML files containing bounding boxes in PASCAL VOC format/val: 94,959 XML files containing bounding boxes in PASCAL VOC format
- PubTables-1M-Image_Table_Words_JSON.tar.gz: Bounding boxes and text content for all of the words in each cropped table image
- One JSON file per cropped table image (plus some extra unused files)
- PubTables-1M-PDF_Annotations_JSON.tar.gz: Detailed annotations for all of the tables appearing in the source PubMed PDFs. All annotations are in PDF coordinates.
- 401,733 JSON files; one file per source PDF
To download from the command line:
- Visit the dataset home page with a web browser and click Download in the top left corner. This will create a link to download the dataset from Azure with a unique access token for you that looks like
https://msropendataset01.blob.core.windows.net/pubtables1m?[SAS_TOKEN_HERE]. - You can then use the command line tool azcopy to download all of the files with the following command:
azcopy copy "https://msropendataset01.blob.core.windows.net/pubtables1m?[SAS_TOKEN_HERE]" "/path/to/your/download/folder/" --recursive
Then unzip each of the archives from the command line using:
tar -xzvf yourfile.tar.gz
Create a conda environment from the yml file and activate it as follows
conda env create -f environment.yml
conda activate tables-detr
The code trains models for 2 different sets of table extraction tasks:
- Table Detection
- Table Structure Recognition + Functional Analysis
For a detailed description of these tasks and the models, please refer to the paper.
To train, you need to cd to the src directory and specify: 1. the path to the dataset, 2. the task (detection or structure), and 3. the path to the config file, which contains the hyperparameters for the architecture and training.
To train the detection model:
python main.py --data_type detection --config_file detection_config.json --data_root_dir /path/to/detection_data
To train the structure recognition model:
python main.py --data_type structure --config_file structure_config.json --data_root_dir /path/to/structure_data
The evaluation code computes standard object detection metrics (AP, AP50, etc.) for both the detection model and the structure model. When running evaluation for the structure model it also computes grid table similarity (GriTS) metrics for table structure recognition. GriTS is a measure of table cell correctness and is defined as the average correctness of each cell averaged over all tables. GriTS can measure the correctness of predicted cells based on: 1. cell topology alone, 2. cell topology and the reported bounding box location of each cell, or 3. cell topology and the reported text content of each cell. For more details on GriTS, please see our papers.
To compute object detection metrics for the detection model:
python main.py --mode eval --data_type detection --config_file detection_config.json --data_root_dir /path/to/pascal_voc_detection_data --model_load_path /path/to/detection_model
To compute object detection and GriTS metrics for the structure recognition model:
python main.py --mode eval --data_type structure --config_file structure_config.json --data_root_dir /path/to/pascal_voc_structure_data --model_load_path /path/to/structure_model --table_words_dir /path/to/json_table_words_data
Optionally you can add flags for things like controlling parallelization, saving detailed metrics, and saving visualizations:
--device cpu: Change the default device from cuda to cpu.
--batch_size 4: Control the batch size to use during the forward pass of the model.
--eval_pool_size 4: Control the worker pool size for CPU parallelization during GriTS metric computation.
--eval_step 2: Control the number of batches of processed input data to accumulate before passing all samples to the parallelized worker pool for GriTS metric computation.
--debug: Create and save visualizations of the model inference. For each input image "PMC1234567_table_0.jpg", this will save two visualizations: "PMC1234567_table_0_bboxes.jpg" containing the bounding boxes output by the model, and "PMC1234567_table_0_cells.jpg" containing the final table cell bounding boxes after post-processing. By default these are saved to a new folder "debug" in the current directory.
--debug_save_dir /path/to/folder: Specify the folder to save visualizations to.
--test_max_size 500: Run evaluation on a randomly sampled subset of the data. Useful for quick verifications and checks.
If model training is interrupted, it can be easily resumed by using the flag --model_load_path /path/to/model.pth and specifying the path to the saved dictionary file that contains the saved optimizer state.
If you want to restart training by fine-tuning a saved checkpoint, such as model_20.pth, use the flag --model_load_path /path/to/model_20.pth and the flag --load_weights_only to indicate that the previous optimizer state is not needed for resuming training.
Whether fine-tuning or training a new model from scratch, you can optionally create a new config file with different training parameters than the default ones we used. Specify the new config file using: --config_file /path/to/new_structure_config.json. Creating a new config file is useful, for example, if you want to use a different learning rate lr during fine-tuning.
Alternatively, many of the arguments in the config file can be specified as command line arguments using their associated flags. Any argument specified as a command line argument overrides the value of the argument in the config file.
Our work can be cited using:
@software{smock2021tabletransformer,
author = {Smock, Brandon and Pesala, Rohith},
month = {06},
title = {{Table Transformer}},
url = {https://github.com/microsoft/table-transformer},
version = {1.0.0},
year = {2021}
}
@inproceedings{smock2022pubtables,
title={Pub{T}ables-1{M}: Towards comprehensive table extraction from unstructured documents},
author={Smock, Brandon and Pesala, Rohith and Abraham, Robin},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)},
pages={4634-4642},
year={2022},
month={June}
}
@inproceedings{smock2023grits,
title={Gri{TS}: Grid table similarity metric for table structure recognition},
author={Smock, Brandon and Pesala, Rohith and Abraham, Robin},
booktitle={International Conference on Document Analysis and Recognition},
pages={535--549},
year={2023},
organization={Springer}
}
@article{smock2023aligning,
title={Aligning benchmark datasets for table structure recognition},
author={Smock, Brandon and Pesala, Rohith and Abraham, Robin},
booktitle={International Conference on Document Analysis and Recognition},
pages={371--386},
year={2023},
organization={Springer}
}
This project welcomes contributions and suggestions. Most contributions require you to agree to a Contributor License Agreement (CLA) declaring that you have the right to, and actually do, grant us the rights to use your contribution. For details, visit https://cla.opensource.microsoft.com.
When you submit a pull request, a CLA bot will automatically determine whether you need to provide a CLA and decorate the PR appropriately (e.g., status check, comment). Simply follow the instructions provided by the bot. You will only need to do this once across all repos using our CLA.
This project has adopted the Microsoft Open Source Code of Conduct. For more information see the Code of Conduct FAQ or contact [email protected] with any additional questions or comments.
This project may contain trademarks or logos for projects, products, or services. Authorized use of Microsoft trademarks or logos is subject to and must follow Microsoft's Trademark & Brand Guidelines. Use of Microsoft trademarks or logos in modified versions of this project must not cause confusion or imply Microsoft sponsorship. Any use of third-party trademarks or logos are subject to those third-party's policies.