philipperemy / keras-tcn Goto Github PK
View Code? Open in Web Editor NEWKeras Temporal Convolutional Network.
License: MIT License
Keras Temporal Convolutional Network.
License: MIT License
Problem:
tensorflow.python.framework.errors_impl.UnknownError: 2 root error(s) found.
(0) Unknown: Failed to get convolution algorithm. This is probably because cuDNN failed to initialize, so try looking to see if a warning log message was printed above.
[[{{node conv1d_1/convolution}}]]
The problem may be because I have got two GPUs (RTX 2080). I ran into the issue with CUDA 10.0 and 10.1
Fix:
Add the following lines to your main.py
The fix has been tested with the "adding problem"
from keras.backend.tensorflow_backend import set_session
import tensorflow as tf
config = tf.ConfigProto()
config.gpu_options.allow_growth = True # dynamically grow the memory used on the GPU
sess = tf.Session(config=config)
I really like this implementation : ). I pulled this down and took the time to understand it. I added a bunch of comments to the functions, do you have any interest in me opening a pull request?
I have a list of sequences as my input data, each sequence could have different time step size, at each time step the feature vector length is 100. I'm using fit_generator in keras to train the model, and a generator function to produce the input with batch_size I specified, so each batch from the generator is an numpy array with shape (batch_size, ), the sub array will have shape (variable_length, 100)
Since my input has different time step size, I specified the input shape using:
Input(batch_shape=(batch_size, timesteps, input_dim))
but this would give me error below:
ValueError: Error when checking input: expected num_input to have 3 dimensions, but got array with shape (128, 1)
where 128 is my batch_size, same error if I set Input(shape=(None, input_dim))
Apologize if this is a dumb question but I am having a problem doing binary classification:
model = compiled_tcn(return_sequences=False,
num_feat=X_train.shape[2],
num_classes=2,
nb_filters=24,
kernel_size=8,
dilations=[2 ** i for i in range(9)],
nb_stacks=1,
max_len=X_train.shape[1],
use_skip_connections=True,
)
model.fit(X_train, y_train, validation_data=(X_val, y_val), epochs=5)
Results in prediction:
model.predict(X_val)
array([[0.5, 0.5],
[0.5, 0.5],
[0.5, 0.5],
[0.5, 0.5],
...,
[0.5, 0.5],
[0.5, 0.5],
[0.5, 0.5],
[0.5, 0.5]], dtype=float32)
Thanks for any help
Input dimension is 30K rows of 83 features, target is 0 or 1
I'm reading the code and I'm just a little confused about the below lines:
for s in range(nb_stacks):
for i in dilatations:
x, skip_out = residual_block(x, s, i, activation, nb_filters, kernel_size)
What is the interpretation of this? In the paper, I had thought that each layer would have its own dilation rate, typically 2 ** i for the ith layer. What is happening here?
I noticed some differences in the implementation of the architecture compared to the original paper.
I've already come across another issue, where @philipperemy was answering: "As the author of the paper stated, this TCN architecture really aims to be a simple baseline that you can improve upon. So personally I think replicating the exact same results is not that important.".
I just wanted to know if there are any reasons behind these two implementation choices that I don't understand:
In the paper, they don't talk about stacking various dilated convolutions (parameter nb_stacks):
I think I understood what you discussed here, but how this changes instead of just increasing the number of dilations?
In the single residual block (in residual_block()), why is there only one convolutional layer instead of 2 as in the paper?
It's just a simplification?
The receptive field is a product of number of stacks, the largest dilation value, and kernel size. Let us say we fix kernel_size = 2 . I would like to understand the difference between two different ways of achieving the same receptive field, say a receptive field of 256:
nb_stacks=1 stacks, and use dilations [1, 2, 4, 8, 16, 32, 64, 128]nb_stacks=2 stacks, and use dilations [1, 2, 4, 8, 16, 32, 64]In general, what kind of tradeoff am I making in choosing to increase stacks versus increase dilations to get the receptive field I want, or are they equivalent?
Can this be used with tensorflow.keras, i.e., can I use the usual keras -> tensorflow.keras substitution?
Your conv_1d setting is:
atrous_rate=2 ** i, i is from [1, 2, 4, 8]
That means you want to make the dilatations of conv1d as
atrous_rate=[2, 4, 16, 256]
My sequence is 100 time-steps. As my guessing, the dilation_rate over 100 cannot be fully effective. But I tried:
atrous_rate=2 ** i, i is from [1, 2, 3, 4, 5, 6]
or
atrous_rate=2 ** i, i is from [1, 2, 4, 6]
Only your configuration with even dilate_rate=256 (>100) gives me the best result.
Can you explain more about this configuration? Thanks!
In tcn/tcn.py
def residual_block(x, dilation_rate, nb_filters, kernel_size, padding, dropout_rate=0):
prev_x = x
for k in range(2):
x = Conv1D(filters=nb_filters,
kernel_size=kernel_size,
dilation_rate=dilation_rate,
padding=padding)(x)
x = Activation('relu')(x)
x = SpatialDropout1D(rate=dropout_rate)(x)
x = Convolution1D(nb_filters, 1, padding='same')(x)
res_x = keras.layers.add([prev_x, x])
return res_x, x
I think the code x = Convolution1D(nb_filters, 1, padding='same')(x) in the penultimate line 3 is not correct. This code want to match the shapes of prev_x and x. I think it's input should be prev_x and not x.
Change the last 3 lines to:
prev_x = Conv1D(nb_filters, 1, padding='same')(prev_x)
res_x = keras.layers.add([prev_x, x])
return res_x, x
` ``
I tried the following things:
`# Set seed value
seed_value = 56
import os
os.environ['PYTHONHASHSEED']=str(seed_value)
python built-in pseudo-random generator at a fixed valueimport random
random.seed(seed_value)
numpy pseudo-random generator at a fixed valueimport numpy as np
np.random.seed(seed_value)
from comet_ml import Experiment
tensorflow pseudo-random generator at a fixed valueimport tensorflow as tf
tf.set_random_seed(seed_value)
tensorflow sessionfrom keras import backend as K
session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
K.set_session(sess)`
However, I cannot still get the same answer by running several times.
Thank you very much!
Best
When using (e.g.)
tcn = TCN(27,dilations=[1, 1, 1, 2, 4, 8, 16, 32],return_sequences=False)(in1)
The code gives an error
"The name "tcn_d_causal_conv_1_tanh_s0" is used 3 times in the model. All layer names should be unique."
This is due to the block naming in 'residual_block' not including the layer number, but only the parameters.
It seems like that channel_normalization layer is different from the weight norm layer in the paper “An Empirical Evaluation of Generic Convolutional and Recurrent Networks for Sequence Modeling”.
Hi,
In your examples the dilations are dilatations=[1, 2, 4, 8]. This is in fact in-line with the imageof the example from WaveNet.
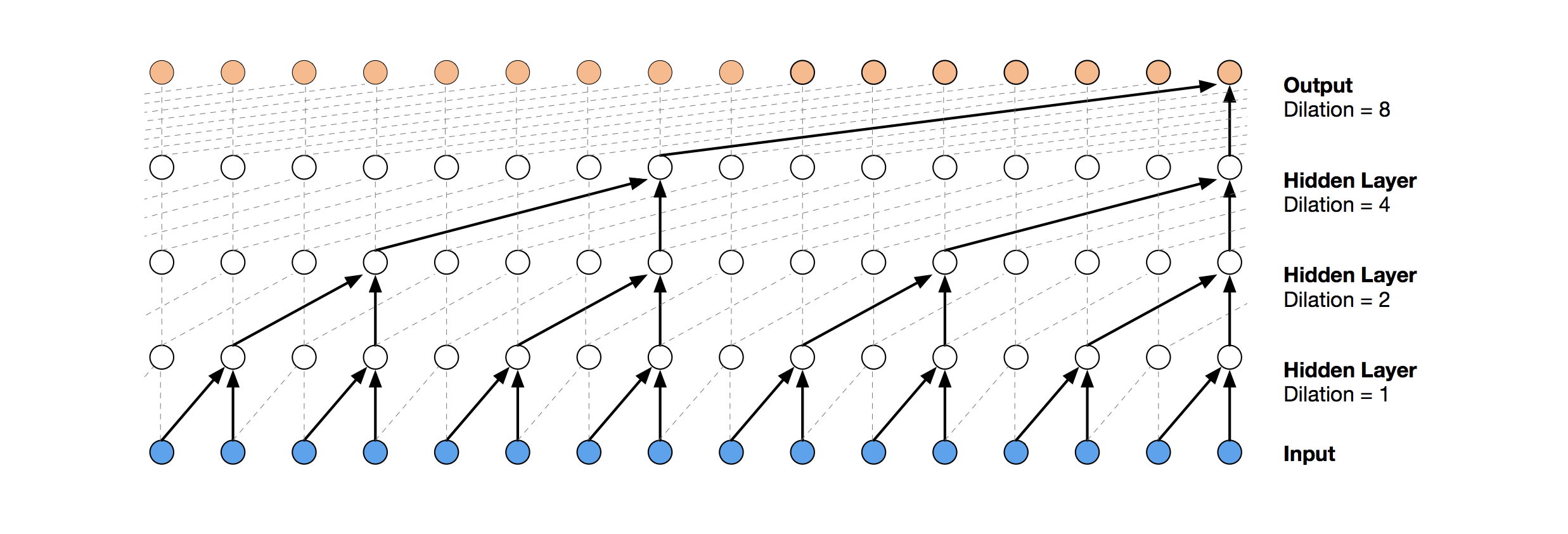
However this will lead to a dilation rate of 2**dilations, which implies the first dilation is 2 instead of 1. Is this a mistake? is there any reason why a dilation_depth parameter is not used instead?
Hi all,
I think there is an issue in copy_memory example
It is giving below error without any change to the code:
InvalidArgumentError: Incompatible shapes: [158976] vs. [256,621]
I could install tcn using pip install keras-tcn successfully without any error.
But when I use it as
"from tcn import tcn" the following error gets generated.
File "/Users/sowmyar/anaconda/lib/python2.7/site-packages/tcn/tcn.py", line 95
print(f'Updated dilations from {dilations} to {new_dilations} because of backwards compatibility.')
^
SyntaxError: invalid syntax
Can you help me with this issue?
Are there plans to support TensorFlow 2.0? When I try to use keras-tnn with TF 2.0, I get this error. Which is likely related to the eager execution model of TF 2.0.
AttributeError Traceback (most recent call last)
in
19 i = Input(batch_shape=(batch_size, timesteps, input_dim))
20
---> 21 o = TCN(return_sequences=False)(i) # The TCN layers are here.
22 o = Dense(1)(o)
23
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/tcn/tcn.py in call(self, inputs)
119 x = inputs
120 # 1D FCN.
--> 121 x = Convolution1D(self.nb_filters, 1, padding=self.padding)(x)
122 skip_connections = []
123 for s in range(self.nb_stacks):
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/keras/legacy/interfaces.py in wrapper(*args, **kwargs)
89 warnings.warn('Update your ' + object_name + ' call to the ' +
90 'Keras 2 API: ' + signature, stacklevel=2)
---> 91 return func(*args, **kwargs)
92 wrapper._original_function = func
93 return wrapper
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/keras/layers/convolutional.py in init(self, filters, kernel_size, strides, padding, data_format, dilation_rate, activation, use_bias, kernel_initializer, bias_initializer, kernel_regularizer, bias_regularizer, activity_regularizer, kernel_constraint, bias_constraint, **kwargs)
357 kernel_constraint=kernel_constraint,
358 bias_constraint=bias_constraint,
--> 359 **kwargs)
360
361 def get_config(self):
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/keras/layers/convolutional.py in init(self, rank, filters, kernel_size, strides, padding, data_format, dilation_rate, activation, use_bias, kernel_initializer, bias_initializer, kernel_regularizer, bias_regularizer, activity_regularizer, kernel_constraint, bias_constraint, **kwargs)
103 bias_constraint=None,
104 **kwargs):
--> 105 super(_Conv, self).init(**kwargs)
106 self.rank = rank
107 self.filters = filters
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/keras/engine/base_layer.py in init(self, **kwargs)
130 if not name:
131 prefix = self.class.name
--> 132 name = to_snake_case(prefix) + '' + str(K.get_uid(prefix))
133 self.name = name
134
~/miniconda3/envs/tensorflow-2.0/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py in get_uid(prefix)
72 """
73 global _GRAPH_UID_DICTS
---> 74 graph = tf.get_default_graph()
75 if graph not in _GRAPH_UID_DICTS:
76 _GRAPH_UID_DICTS[graph] = defaultdict(int)
AttributeError: module 'tensorflow' has no attribute 'get_default_graph'
`
I tried instantiating TCN via the complied_tcn function in this way:
classifier = compiled_tcn(num_feat=train_X.shape[2],
num_classes=4,
nb_filters=train_X.shape[1],
kernel_size=3*train_X.shape[2],
dilations=[2 ** i for i in range(9)],
nb_stacks=1,
max_len=train_X.shape[1],
use_skip_connections=True,
opt='rmsprop',
lr=5e-4,
regression=False,
return_sequences=False)
This is the output from running the line above:
x.shape= (?, 80)
model.x = (?, 80, 26)
model.y = (?, 4)
Adam with norm clipping.
These are the shapes of my train and test sets:
[in]: print(train_X.shape, train_y.shape, test_X.shape, test_y.shape)
(10540, 80, 26) (10540, 4) (555, 80, 26) (555, 4)
When I attempt to run fit on the classifier with this line:
classifier.fit(train_X, train_y, validation_data=(test_X, test_y), epochs=epochs,batch_size=batch_size)
I get this error:
ValueError: Error when checking target: expected activation_133 to have shape (1,) but got array with shape (4,)
I don't understand where is the shape (1,) coming from, since I have num_classes=4, regression=False and return_sequences=False and the shapes of my data and the model seem to match as well.
Testing the regression example from the README,
model = tcn.dilated_tcn(output_slice_index='last',
num_feat=20,
nb_filters=24,
kernel_size=8,
dilatations=[1, 2, 4, 8],
nb_stacks=8,
max_len=100,
activation='norm_relu',
regression=True)
gives the error
Traceback (most recent call last):
File "<stdin>", line 9, in <module>
TypeError: dilated_tcn() missing 1 required positional argument: 'num_classes'
I'm trying to replicate the result of my previous project that used LSTM with units = 256. And on the home page of this repo it says that number of filters is like units in LSTM. And since I want the tcn to remember the whole sequence I set the receptive field about equals to the sequence length. But with number of filters 256 I ended up with a network that is way larger than the LSTM counterpart. So how do I calculate the appropriate number of filters?
Line 30 in c679925
I can find that in the original paper, the normalization layer comes before the Relu activation layer. Why this isn't the case here?
Thank you!!
I want to learn how to process text data using tf.keras and tcn
Hello i use plaidml as keras backend because it supports AMD graphics cards
there are two issues with your implementation
spatialdropout is not supported by plaidml so
x = SpatialDropout1D(dropout_rate, name='spatial_dropout1d_%d_s%d_%f' % (i, s, dropout_rate))(x)
will not work
plaidml/plaidml#165
it seems that keras.backend.max isn't implemented too
If you don't have time to fix it can you suggest a workaround?
"dilations" is treated as an optional argument that defaults to "None": tcn.TCN(nb_filters=64, kernel_size=2, nb_stacks=1, dilations=None, activation='norm_relu', padding='causal', use_skip_connections=True, dropout_rate=0.0, return_sequences=True, name='tcn')
In the readme, it only states "List. A dilation list. Example is: [1, 2, 4, 8, 16, 32, 64]."
and in the __call__ function of tcn.py it seams that it gets converted to [1, 2, 4, 8, 16, 32, 64] when it's set to None.
In this case, I think it it should either default to [1, 2, 4, 8, 16, 32] directly instead of None or it should at least be stated in the docs that it behaves so since it's quite confusing this way. Or did i miss something?
As mentioned in title.
Somewhere you seem to have forgotten to use the activation function
Hi, thanks for offering this great package. I'm trying to build a autoencoder using TCN. In my encoder part, the TCN will not return a sequence. After looking at your code, it seems to me that if return sequence is set to False, then only one slice in the last layer is kept for output. However, the convolution is still conducted for other slices. Tracing back the dilated convolution, many computations that only affect the discarded slices is wasted. I'm not sure if I understand it correctly. If not, please kindly point out.
I also have a question in building the decoder. I'm not sure if I should use a identical dilation order in the TCN group of layers as in the encoder, or should reverse the dilation order. This is a general question not very related to the implementation of TCN itself so you can ignore it if you'd like to. Any comment/suggestions would be appreciated.
The name parameter is currently not being applied to the layer. As a related issue, the current layer implementation dumps all of the model layers into a single flattened list and it's hard to see where one TCN block ends and another begins. I propose that, to solve both of these issues, the TCN layer should inherit from keras.layers.Layer
I patched the layer to implement the desired interface here, but all of this can be factored into the original TCN layer.
from keras.layers import Layer
class TCN2(Layer):
def __init__(self, *a, name=None, **kw):
self.model = TCN(*a, **kw)
Layer.__init__(self, name=name)
def call(self, x):
return self.model(x)
def compute_output_shape(self, input_shape):
return input_shape[:-1] + (self.model.nb_filters,)Here's a sketch of the changes translated to the original class:
from keras.layers import Layer
class TCN(Layer):
# remove name from args here so it is passed in **kw
def __init__(self, *a, ..., **kw):
...
super().__init__(*a, **kw)
call = __call__ # just change the method name
def compute_output_shape(self, input_shape):
return input_shape[:-1] + (self.nb_filters,)Now when I do this:
batch_size, timesteps, input_ch = None, fs*dur, 1
i = Input(batch_shape=(batch_size, timesteps, input_ch), name='tcn_in')
o = TCN2(16, return_sequences=True, name='tcn1')(i)
o = TCN2(16, return_sequences=True, name='tcn2')(o)
o = TCN2(16, return_sequences=True, name='tcn3')(o)
m = Model(inputs=[i], outputs=[o])
m.compile(optimizer='adam', loss='mse')
[l.name for l in m.layers]I get:
['tcn_in', 'tcn1', 'tcn2', 'tcn3']
Instead of:
['tcn_in',
'conv1d_927',
'conv1d_928',
'activation_871',
'spatial_dropout1d_581',
'conv1d_929',
'activation_872',
'spatial_dropout1d_582',
'conv1d_930',
'add_334',
'activation_873',
'conv1d_931',
'activation_874',
'spatial_dropout1d_583',
'conv1d_932',
'activation_875',
'spatial_dropout1d_584',
'conv1d_933',
'add_335',
'activation_876',
'conv1d_934',
'activation_877',
'spatial_dropout1d_585',
'conv1d_935',
'activation_878',
'spatial_dropout1d_586',
'conv1d_936',
'add_336',
'activation_879',
'conv1d_937',
'activation_880',
'spatial_dropout1d_587',
'conv1d_938',
'activation_881',
'spatial_dropout1d_588',
'conv1d_939',
'add_337',
'activation_882',
'conv1d_940',
'activation_883',
'spatial_dropout1d_589',
'conv1d_941',
'activation_884',
'spatial_dropout1d_590',
'conv1d_942',
'add_338',
'activation_885',
'conv1d_943',
'activation_886',
'spatial_dropout1d_591',
'conv1d_944',
'activation_887',
'spatial_dropout1d_592',
'add_340',
'conv1d_946',
'conv1d_947',
'activation_889',
'spatial_dropout1d_593',
'conv1d_948',
'activation_890',
'spatial_dropout1d_594',
'conv1d_949',
'add_341',
'activation_891',
'conv1d_950',
'activation_892',
'spatial_dropout1d_595',
'conv1d_951',
'activation_893',
'spatial_dropout1d_596',
'conv1d_952',
'add_342',
'activation_894',
'conv1d_953',
'activation_895',
'spatial_dropout1d_597',
'conv1d_954',
'activation_896',
'spatial_dropout1d_598',
'conv1d_955',
'add_343',
'activation_897',
'conv1d_956',
'activation_898',
'spatial_dropout1d_599',
'conv1d_957',
'activation_899',
'spatial_dropout1d_600',
'conv1d_958',
'add_344',
'activation_900',
'conv1d_959',
'activation_901',
'spatial_dropout1d_601',
'conv1d_960',
'activation_902',
'spatial_dropout1d_602',
'conv1d_961',
'add_345',
'activation_903',
'conv1d_962',
'activation_904',
'spatial_dropout1d_603',
'conv1d_963',
'activation_905',
'spatial_dropout1d_604',
'add_347',
'conv1d_965',
'conv1d_966',
'activation_907',
'spatial_dropout1d_605',
'conv1d_967',
'activation_908',
'spatial_dropout1d_606',
'conv1d_968',
'add_348',
'activation_909',
'conv1d_969',
'activation_910',
'spatial_dropout1d_607',
'conv1d_970',
'activation_911',
'spatial_dropout1d_608',
'conv1d_971',
'add_349',
'activation_912',
'conv1d_972',
'activation_913',
'spatial_dropout1d_609',
'conv1d_973',
'activation_914',
'spatial_dropout1d_610',
'conv1d_974',
'add_350',
'activation_915',
'conv1d_975',
'activation_916',
'spatial_dropout1d_611',
'conv1d_976',
'activation_917',
'spatial_dropout1d_612',
'conv1d_977',
'add_351',
'activation_918',
'conv1d_978',
'activation_919',
'spatial_dropout1d_613',
'conv1d_979',
'activation_920',
'spatial_dropout1d_614',
'conv1d_980',
'add_352',
'activation_921',
'conv1d_981',
'activation_922',
'spatial_dropout1d_615',
'conv1d_982',
'activation_923',
'spatial_dropout1d_616',
'add_354']
The default weight initialization in Keras is Glorot Uniform. I've read that for Conv2D layers you should use the initialization algorithm from Kaiming He instead. I'm not sure if that applies to 1D Conv that are used in keras-tcn, but it may be nice to have the choice regardless.
Can this be used as a layer in an autoencoder by stacking the TCN layers? Or would you need to do UpSampling1D ?
Hi Philip,
I am using keras-tcn and it works really well. Thanks for your contribution.
I can save the weights of my model using:
ModelCheckpoint('weights.h5', save_weights_only=True)
However, if I use:
model.save_weights('weights.h5')
I get the following error:
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _do_call(self, fn, *args)
1333 try:
-> 1334 return fn(*args)
1335 except errors.OpError as e:
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _run_fn(feed_dict, fetch_list, target_list, options, run_metadata)
1318 return self._call_tf_sessionrun(
-> 1319 options, feed_dict, fetch_list, target_list, run_metadata)
1320
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _call_tf_sessionrun(self, options, feed_dict, fetch_list, target_list, run_metadata)
1406 self._session, options, feed_dict, fetch_list, target_list,
-> 1407 run_metadata)
1408
FailedPreconditionError: Attempting to use uninitialized value conv1d_33/bias
[[{{node conv1d_33/bias/_0}} = _Send[T=DT_FLOAT, client_terminated=false, recv_device="/job:localhost/replica:0/task:0/device:CPU:0", send_device="/job:localhost/replica:0/task:0/device:GPU:0", send_device_incarnation=1, tensor_name="edge_134_conv1d_33/bias", _device="/job:localhost/replica:0/task:0/device:GPU:0"](conv1d_33/bias)]]
[[{{node tcn_d_same_conv_4_tanh_s0_2/kernel/_99}} = _Recv[_start_time=0, client_terminated=false, recv_device="/job:localhost/replica:0/task:0/device:CPU:0", send_device="/job:localhost/replica:0/task:0/device:GPU:0", send_device_incarnation=1, tensor_name="edge_232_tcn_d_same_conv_4_tanh_s0_2/kernel", tensor_type=DT_FLOAT, _device="/job:localhost/replica:0/task:0/device:CPU:0"]()]]
During handling of the above exception, another exception occurred:
FailedPreconditionError Traceback (most recent call last)
<ipython-input-31-f4a33930d8c0> in <module>()
18 PD_signals=PD_signals, nonPD_same=nonPD_same, nonPD_different=nonPD_different, sig_shape=sig_shape)
19
---> 20 model.save_weights(models_dir + name + '_weights.h5') # creates a HDF5 file 'my_model.h5'
21
22 # ModelCheckpoint(models_dir + name + '_weights.h5', save_weights_only=True)
~/anaconda2/envs/py35/lib/python3.5/site-packages/keras/engine/network.py in save_weights(self, filepath, overwrite)
1119 return
1120 with h5py.File(filepath, 'w') as f:
-> 1121 saving.save_weights_to_hdf5_group(f, self.layers)
1122 f.flush()
1123
~/anaconda2/envs/py35/lib/python3.5/site-packages/keras/engine/saving.py in save_weights_to_hdf5_group(f, layers)
570 g = f.create_group(layer.name)
571 symbolic_weights = layer.weights
--> 572 weight_values = K.batch_get_value(symbolic_weights)
573 weight_names = []
574 for i, (w, val) in enumerate(zip(symbolic_weights, weight_values)):
~/anaconda2/envs/py35/lib/python3.5/site-packages/keras/backend/tensorflow_backend.py in batch_get_value(ops)
2418 """
2419 if ops:
-> 2420 return get_session().run(ops)
2421 else:
2422 return []
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in run(self, fetches, feed_dict, options, run_metadata)
927 try:
928 result = self._run(None, fetches, feed_dict, options_ptr,
--> 929 run_metadata_ptr)
930 if run_metadata:
931 proto_data = tf_session.TF_GetBuffer(run_metadata_ptr)
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _run(self, handle, fetches, feed_dict, options, run_metadata)
1150 if final_fetches or final_targets or (handle and feed_dict_tensor):
1151 results = self._do_run(handle, final_targets, final_fetches,
-> 1152 feed_dict_tensor, options, run_metadata)
1153 else:
1154 results = []
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _do_run(self, handle, target_list, fetch_list, feed_dict, options, run_metadata)
1326 if handle is None:
1327 return self._do_call(_run_fn, feeds, fetches, targets, options,
-> 1328 run_metadata)
1329 else:
1330 return self._do_call(_prun_fn, handle, feeds, fetches)
~/anaconda2/envs/py35/lib/python3.5/site-packages/tensorflow/python/client/session.py in _do_call(self, fn, *args)
1346 pass
1347 message = error_interpolation.interpolate(message, self._graph)
-> 1348 raise type(e)(node_def, op, message)
1349
1350 def _extend_graph(self):
FailedPreconditionError: Attempting to use uninitialized value conv1d_33/bias
[[{{node conv1d_33/bias/_0}} = _Send[T=DT_FLOAT, client_terminated=false, recv_device="/job:localhost/replica:0/task:0/device:CPU:0", send_device="/job:localhost/replica:0/task:0/device:GPU:0", send_device_incarnation=1, tensor_name="edge_134_conv1d_33/bias", _device="/job:localhost/replica:0/task:0/device:GPU:0"](conv1d_33/bias)]]
[[{{node tcn_d_same_conv_4_tanh_s0_2/kernel/_99}} = _Recv[_start_time=0, client_terminated=false, recv_device="/job:localhost/replica:0/task:0/device:CPU:0", send_device="/job:localhost/replica:0/task:0/device:GPU:0", send_device_incarnation=1, tensor_name="edge_232_tcn_d_same_conv_4_tanh_s0_2/kernel", tensor_type=DT_FLOAT, _device="/job:localhost/replica:0/task:0/device:CPU:0"]()]]
It seems as if "conv1d_33" (which is part of the TCN in my model) is considered as "uninitialized" by the method "save_model". Do you have any suggestion why this could be the case?
Thanks again for you great library
Hi,
Which versions of Tensorflow and Keras was this tested on? I keep getting issues that I think are related to incompatible version.
Hi Phillippe, thanks for doing this code, it is very helpful and works well.
I am doing time series prediction so can choose a sequence as long or as short as I want for the TCN to work with. What I am not clear on is how the kernel size, dilation values and number of stacks interact to determine the receptive field size.
For example, if I have a kernel size=8, dilation values of [1,2,4,8] and stacks=3, how long should my sequence be so that it fully covers the receptive field of the TCN? Can you clarify?
I'm trying to extract context vectors from the latent TCN space. Basically I want to extract all of the highlighted nodes from the hidden layers in this diagram (nodes with arrows) into a list of activations, one for each residual block:
[
np.array(shape=(8, n_ch)), # finest temporal activations
np.array(shape=(4, n_ch)), # middle temporal activations
np.array(shape=(2, n_ch)), # coarsest temporal activations
]The way that I see it, I need to get the activation layers at the end of each residual block, get their output tensors, and slice them using a stride based on the dilation rate. I'm just not sure if I need to do anything special to handle dilation rates of stacked TCN blocks. I think it would be helpful to add a helper method/property that makes it easier to extract this context.
If you're wondering about my use case, I'm trying to build a time series autoencoder-style network for multi-scale anomaly detection and I'm trying to model the learned latent space at multiple temporal scales using some distribution (probably a gaussian mixture model atm).
I would like to expand the code with tf.keras. Can I modify the code to release it on pypi?
I know the license is MIT, but I would like to confirm that just in case.
Hi,
The shape of my input data is x_train: (856,119,68), y_train: (856,1)
where 68 are the dimension of the feature vector. I define model like this
model = tcn.compiled_tcn(num_feat=68,
num_classes=5,
nb_filters=48,
kernel_size=68,
dilations=[1, 2, 4], #[2 ** i for i in range(9)],
nb_stacks=2,
max_len=119, #train_x[0:1].shape[1],
activation='norm_relu',
use_skip_connections=True,
return_sequences=True)
When I run
model.fit(train_x, train_y, validation_data=(test_x, test_y), epochs=100,
batch_size=256)
I get this following error
"""Error when checking target: expected activation_59 to have 3 dimensions, but got array with shape (856, 1)"""
Please let me know if I have to change anything in the code?
Running
from tcn import tcn
model = tcn.dilated_tcn(output_slice_index='last',
num_feat=20,
num_classes=None,
nb_filters=24,
kernel_size=8,
dilatations=[1, 2, 4, 8],
nb_stacks=8,
max_len=100,
activation='norm_relu',
regression=True)
gives the Warnings
Using TensorFlow backend.
/home/hoppeta/.pyenv/versions/3.6.0/lib/python3.6/site-packages/keras/legacy/layers.py:748: UserWarning: The `AtrousConvolution1D` layer has been deprecated. Use instead the `Conv1D` layer with the `dilation_rate` argument.
warnings.warn('The `AtrousConvolution1D` layer '
/home/hoppeta/src/keras-tcn/tcn/tcn.py:38: UserWarning: The `Merge` layer is deprecated and will be removed after 08/2017. Use instead layers from `keras.layers.merge`, e.g. `add`, `concatenate`, etc.
res_x = Merge(mode='sum')([original_x, x])
/home/hoppeta/src/keras-tcn/tcn/tcn.py:62: UserWarning: The `Merge` layer is deprecated and will be removed after 08/2017. Use instead layers from `keras.layers.merge`, e.g. `add`, `concatenate`, etc.
x = Merge(mode='sum')(skip_connections)
x.shape= (?, 24)
model.x = (?, 100, 20)
model.y = (?, 1)
Hi, first of all, thanks for your code sharing.
Guessing your intentions, the following expression may be appropriate.
def process_dilations(dilations):
...
else:
new_dilations = [2 ** i for i in dilations]
--> new_dilations = [2 ** i for i in range(len(dilations))]
Hi philipperemy,
After I update tcn to the recent version, the training accuracy reduced tremendously (around from 0.98 to 0.17)
I then uncommented the following line of the code for BatchNormalization and things seems to be back to normal. So this seems to be an important part of tcn
x = BatchNormalization()(x) # TODO should be WeightNorm here.
Is there a quick fix that going to happen soon?
Thank you
This comment in tcn.py doesn't seem to jive with the code:
num_feat: A tensor of shape (batch_size, timesteps, input_dim).
Thanks for creating this easy to access library. I am using it for time series forecasting scenario based on the example given. After training the model, I saved it in h5 format using save method in Keras. However, when I try to load the model again using load_model() it gives an error saying
This version performs the same function as Dropout, however it drops entire 1D feature maps instead of individual elements. If adjacent frames within feature maps are strongly correlated (as is normally the case in early convolution layers) then regular dropout will not regularize the
IndexError: tuple index out of range
After some google search, I figured it might be because of the Lambda layer used in the model. In this scenario, what is the best option to save and load the TCN model ?
Hello everyone,
we'd like to try your TCN layers for speech enhancement task.
Our model is an autoencoder which takes ~270ms of spectrogram data (noisy speech)with dimensionality (F, T, C) where:
It is in our understanding that we have to:
However, it is not clear how to obtain the original dimensionality after stacking one or more TCNs, as well as how to the layer in a way that reduces/expands the dimensionality of the data.
Could you shed some light on it and/or highlight any pitfall in our approach?
Thanks in advance!
Hello,
I have to apologize that I'm not a math expert, but I wish to implement a GAN network using TCN in the discriminator model to process a generated sequence with dynamic length. Is it possible to make TCN stateful so it can process dynamic length data?
And if yes, is it possible to integrate stateful TCN to seq2seq model?
Thanks so much and I'm very appreciated to you and your project,
Tobias.
I have tried to use your library to do a many to many regression, which does not seem to be supported out of the box. Would you mind to indicate what has to be changed for this use case?
How to input the feature vector of voice data into TCN? Thank you.
I did not modify any parameters, but I got a val_accuracy about 0.11 till many epochs:
55000/55000 [==============================] - 164s 3ms/step - loss: 2.3037 - accuracy: 0.1120 - val_loss: 2.3015 - val_accuracy: 0.1135
Is there any special setting in this example("mnist_pixel")?
ps: I try other examples too, and the result seems good.
I refer to TemporalBlock in https://github.com/locuslab/TCN/blob/master/TCN/tcn.py
I found some slight difference in code https://github.com/philipperemy/keras-tcn/blob/master/tcn/tcn.py.
Line 131:
if use_skip_connections:
x = keras.layers.add(skip_connections)
i think it should be:
if use_skip_connections: x = keras.layers.add([skip_connections,x])
if i am wrong , pls correct me . Thanks
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.