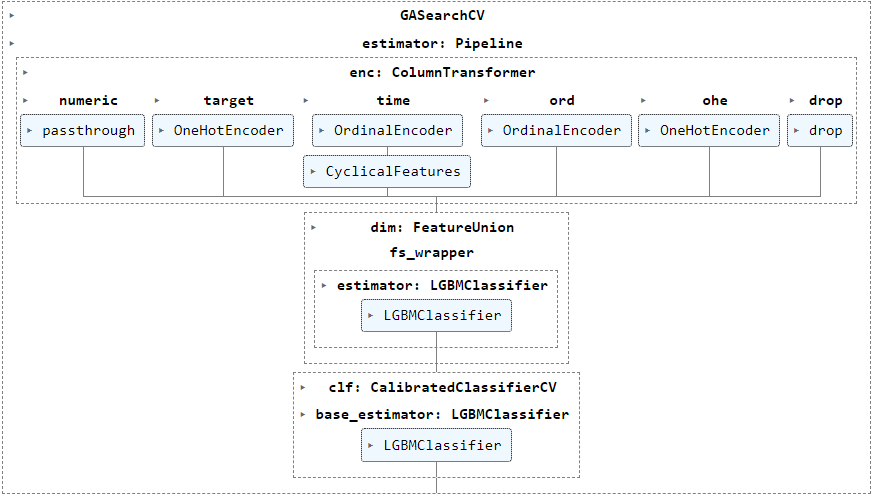
Has anyone come across the issue described below? I'd appreciate any direction to help resolve this.
{'algorithm': 'eaMuPlusLambda',
'criteria': 'max',
'crossover_probability': 0.2,
'cv': StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
'elitism': 1,
'error_score': nan,
<<< estimator parameters removed for brevity >>>
'generations': 10,
'keep_top_k': 1,
'log_config': None,
'mutation_probability': 0.8,
'n_jobs': 18,
'param_grid': {'enc__numeric': <sklearn_genetic.space.space.Categorical at 0x25b40d91160>,
'enc__target': <sklearn_genetic.space.space.Categorical at 0x25b40d98520>,
'enc__time__cyclicity': <sklearn_genetic.space.space.Categorical at 0x25b40d981f0>,
'dim__fs_wrapper': <sklearn_genetic.space.space.Categorical at 0x25b40d984c0>,
'clf__base_estimator__max_depth': <sklearn_genetic.space.space.Integer at 0x25b40cf3d90>,
'clf__base_estimator__num_leaves': <sklearn_genetic.space.space.Integer at 0x25b40d98460>,
'clf__base_estimator__min_child_samples': <sklearn_genetic.space.space.Integer at 0x25b40d98220>,
'clf__base_estimator__colsample_bytree': <sklearn_genetic.space.space.Continuous at 0x25b40265c70>,
'clf__base_estimator__subsample': <sklearn_genetic.space.space.Continuous at 0x25b40265d60>,
'clf__base_estimator__learning_rate': <sklearn_genetic.space.space.Continuous at 0x25b40265ca0>,
'clf__base_estimator__min_split_gain': <sklearn_genetic.space.space.Continuous at 0x25b40265fd0>},
'population_size': 10,
'pre_dispatch': '2*n_jobs',
'refit': 'avg_prec',
'return_train_score': True,
'scoring': {'profit_ratio': make_scorer(profit_ratio_score),
'f_0.5': make_scorer(fbeta_score, beta=0.5, zero_division=0, pos_label=1),
'precision': make_scorer(precision_score, zero_division=0, pos_label=1),
'recall': make_scorer(recall_score, zero_division=0, pos_label=1),
'avg_prec': make_scorer(average_precision_score, needs_proba=True, pos_label=1),
'brier_rel': make_scorer(brier_rel_score, needs_proba=True, pos_label=1),
'brier_score': make_scorer(brier_score_loss, greater_is_better=False, needs_proba=True, pos_label=1),
'logloss_rel': make_scorer(logloss_rel_score, needs_proba=True),
'log_loss': make_scorer(log_loss, greater_is_better=False, needs_proba=True)},
'tournament_size': 3,
'verbose': 1}
The following param_grid (generated using BayesSearchCV to show real information instead of the objects) show categorical values for various transformers steps enc__numeric, enc__target, enc__time__cyclicity and dim__fs_wrapper besides numerical parameter ranges for clf__base_estimator.
{'enc__numeric': Categorical(categories=('passthrough',
SmartCorrelatedSelection(selection_method='variance', threshold=0.9),
SmartCorrelatedSelection(cv='skf5',
estimator=LGBMClassifier(learning_rate=1.0,
max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1),
selection_method='model_performance', threshold=0.9)), prior=None),
'enc__target': Categorical(categories=(MeanEncoder(ignore_format=True), TargetEncoder(), MEstimateEncoder(),
WoEEncoder(ignore_format=True), PRatioEncoder(ignore_format=True),
BayesianTargetEncoder(columns=['Symbol', 'CandleType', 'h1CandleType1', 'h2CandleType1'],
prior_weight=3, suffix='')), prior=None),
'enc__time__cyclicity': Categorical(categories=(CyclicalFeatures(drop_original=True),
CycleTransformer(), RepeatingBasisFunction(n_periods=96)), prior=None),
'dim__fs_wrapper': Categorical(categories=('passthrough',
SelectFromModel(estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_'),
RFECV(cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877, n_jobs=1,
num_leaves=59, random_state=0, subsample=0.1,
verbose=-1),
importance_getter='feature_importances_', min_features_to_select=10, n_jobs=1,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1), step=3),
GeneticSelectionCV(caching=True,
cv=StratifiedKFold(n_splits=5, random_state=0, shuffle=True),
estimator=LGBMClassifier(learning_rate=1.0, max_depth=8,
min_child_samples=4,
min_split_gain=0.0031642299495941877,
n_jobs=1, num_leaves=59,
random_state=0, subsample=0.1,
verbose=-1),
mutation_proba=0.1, n_gen_no_change=3, n_generations=20, n_population=50,
scoring=make_scorer(average_precision_score, needs_proba=True, pos_label=1))), prior=None),
'clf__base_estimator__eval_metric': Categorical(categories=('logloss', 'aucpr'), prior=None),
'clf__base_estimator__max_depth': Integer(low=2, high=8, prior='uniform', transform='identity'),
'clf__base_estimator__min_child_weight': Real(low=1e-05, high=1000, prior='log-uniform', transform='identity'),
'clf__base_estimator__colsample_bytree': Real(low=0.1, high=1.0, prior='uniform', transform='identity'),
'clf__base_estimator__subsample': Real(low=0.1, high=0.9999999999999999, prior='uniform', transform='identity'),
'clf__base_estimator__learning_rate': Real(low=1e-05, high=1, prior='log-uniform', transform='identity'),
'clf__base_estimator__gamma': Real(low=1e-06, high=1000, prior='log-uniform', transform='identity')}
But on many occasions, the error occurs after all the generations seem to have run successfully as indicated by the results below:
C:\Anaconda3\envs\py38_skl\lib\site-packages\deap\creator.py:138: RuntimeWarning: A class named 'Individual' has already been created and it will be overwritten. Consider deleting previous creation of that class or rename it.
warnings.warn("A class named '{0}' has already been created and it "
gen nevals fitness fitness_std fitness_max fitness_min
0 10 0.479343 0.0470905 0.567048 0.436592
1 20 0.508531 0.0540786 0.567048 0.436592
2 20 0.544683 0.0443227 0.567048 0.436592
3 20 0.567027 0.000751322 0.567867 0.565522
4 20 0.571346 0.00762421 0.593858 0.567867
5 20 0.573536 0.0101964 0.593858 0.567867
6 20 0.580379 0.0114497 0.593858 0.567867
7 20 0.582027 0.0117464 0.593858 0.567867
8 20 0.593278 0.00492738 0.598567 0.579708
9 20 0.596899 0.0025436 0.599324 0.592604
10 20 0.597076 0.00379484 0.603329 0.592604
AssertionError Traceback (most recent call last)
Cell In [149], line 2
1 start = time()
----> 2 sv_results_gen = cross_val_thresh(gen_pipe, X, y, cv_val[VAL], result_metrics, #scoring=score_metrics,
3 return_estimator=True, return_train_score=True,
4 thresh_split=SPLIT,
5 )
6 end = time()
Cell In [119], line 30, in cross_val_thresh(estimator, X, y, cv, result_metrics, return_estimator, return_train_score, thresh_split, *args, **kwargs)
27 time_df.loc[i, 'split'] = i
29 start = time()
---> 30 est_i.fit(X_train, y_train)
31 end = time()
32 time_df.loc[i, 'fit_time'] = end - start
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn_genetic\genetic_search.py:555, in GASearchCV.fit(self, X, y, callbacks)
552 self.estimator.set_params(**self.best_params_)
554 refit_start_time = time.time()
--> 555 self.estimator.fit(self.X_, self.y_)
556 refit_end_time = time.time()
557 self.refit_time_ = refit_end_time - refit_start_time
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:378, in Pipeline.fit(self, X, y, **fit_params)
352 """Fit the model.
353
354 Fit all the transformers one after the other and transform the
(...)
375 Pipeline with fitted steps.
376 """
377 fit_params_steps = self._check_fit_params(**fit_params)
--> 378 Xt = self._fit(X, y, **fit_params_steps)
379 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)):
380 if self._final_estimator != "passthrough":
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:336, in Pipeline._fit(self, X, y, **fit_params_steps)
334 cloned_transformer = clone(transformer)
335 # Fit or load from cache the current transformer
--> 336 X, fitted_transformer = fit_transform_one_cached(
337 cloned_transformer,
338 X,
339 y,
340 None,
341 message_clsname="Pipeline",
342 message=self._log_message(step_idx),
343 **fit_params_steps[name],
344 )
345 # Replace the transformer of the step with the fitted
346 # transformer. This is necessary when loading the transformer
347 # from the cache.
348 self.steps[step_idx] = (name, fitted_transformer)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\memory.py:594, in MemorizedFunc.call(self, *args, **kwargs)
593 def call(self, *args, **kwargs):
--> 594 return self._cached_call(args, kwargs)[0]
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\memory.py:537, in MemorizedFunc._cached_call(self, args, kwargs, shelving)
534 must_call = True
536 if must_call:
--> 537 out, metadata = self.call(*args, **kwargs)
538 if self.mmap_mode is not None:
539 # Memmap the output at the first call to be consistent with
540 # later calls
541 if self._verbose:
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\memory.py:779, in MemorizedFunc.call(self, *args, **kwargs)
777 if self._verbose > 0:
778 print(format_call(self.func, args, kwargs))
--> 779 output = self.func(*args, **kwargs)
780 self.store_backend.dump_item(
781 [func_id, args_id], output, verbose=self._verbose)
783 duration = time.time() - start_time
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:1154, in FeatureUnion.fit_transform(self, X, y, **fit_params)
1133 def fit_transform(self, X, y=None, **fit_params):
1134 """Fit all transformers, transform the data and concatenate results.
1135
1136 Parameters
(...)
1152 sum of n_components (output dimension) over transformers.
1153 """
-> 1154 results = self._parallel_func(X, y, fit_params, _fit_transform_one)
1155 if not results:
1156 # All transformers are None
1157 return np.zeros((X.shape[0], 0))
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:1176, in FeatureUnion._parallel_func(self, X, y, fit_params, func)
1173 self._validate_transformer_weights()
1174 transformers = list(self._iter())
-> 1176 return Parallel(n_jobs=self.n_jobs)(
1177 delayed(func)(
1178 transformer,
1179 X,
1180 y,
1181 weight,
1182 message_clsname="FeatureUnion",
1183 message=self._log_message(name, idx, len(transformers)),
1184 **fit_params,
1185 )
1186 for idx, (name, transformer, weight) in enumerate(transformers, 1)
1187 )
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\parallel.py:1085, in Parallel.call(self, iterable)
1076 try:
1077 # Only set self._iterating to True if at least a batch
1078 # was dispatched. In particular this covers the edge
(...)
1082 # was very quick and its callback already dispatched all the
1083 # remaining jobs.
1084 self._iterating = False
-> 1085 if self.dispatch_one_batch(iterator):
1086 self._iterating = self._original_iterator is not None
1088 while self.dispatch_one_batch(iterator):
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\parallel.py:901, in Parallel.dispatch_one_batch(self, iterator)
899 return False
900 else:
--> 901 self._dispatch(tasks)
902 return True
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\parallel.py:819, in Parallel._dispatch(self, batch)
817 with self._lock:
818 job_idx = len(self._jobs)
--> 819 job = self._backend.apply_async(batch, callback=cb)
820 # A job can complete so quickly than its callback is
821 # called before we get here, causing self._jobs to
822 # grow. To ensure correct results ordering, .insert is
823 # used (rather than .append) in the following line
824 self._jobs.insert(job_idx, job)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib_parallel_backends.py:208, in SequentialBackend.apply_async(self, func, callback)
206 def apply_async(self, func, callback=None):
207 """Schedule a func to be run"""
--> 208 result = ImmediateResult(func)
209 if callback:
210 callback(result)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib_parallel_backends.py:597, in ImmediateResult.init(self, batch)
594 def init(self, batch):
595 # Don't delay the application, to avoid keeping the input
596 # arguments in memory
--> 597 self.results = batch()
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\parallel.py:288, in BatchedCalls.call(self)
284 def call(self):
285 # Set the default nested backend to self._backend but do not set the
286 # change the default number of processes to -1
287 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 288 return [func(*args, **kwargs)
289 for func, args, kwargs in self.items]
File C:\Anaconda3\envs\py38_skl\lib\site-packages\joblib\parallel.py:288, in (.0)
284 def call(self):
285 # Set the default nested backend to self._backend but do not set the
286 # change the default number of processes to -1
287 with parallel_backend(self._backend, n_jobs=self._n_jobs):
--> 288 return [func(*args, **kwargs)
289 for func, args, kwargs in self.items]
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\utils\fixes.py:117, in _FuncWrapper.call(self, *args, **kwargs)
115 def call(self, *args, **kwargs):
116 with config_context(**self.config):
--> 117 return self.function(*args, **kwargs)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\pipeline.py:870, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params)
868 with _print_elapsed_time(message_clsname, message):
869 if hasattr(transformer, "fit_transform"):
--> 870 res = transformer.fit_transform(X, y, **fit_params)
871 else:
872 res = transformer.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\sklearn\base.py:870, in TransformerMixin.fit_transform(self, X, y, **fit_params)
867 return self.fit(X, **fit_params).transform(X)
868 else:
869 # fit method of arity 2 (supervised transformation)
--> 870 return self.fit(X, y, **fit_params).transform(X)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\genetic_selection\gscv.py:279, in GeneticSelectionCV.fit(self, X, y, groups)
262 def fit(self, X, y, groups=None):
263 """Fit the GeneticSelectionCV model and then the underlying estimator on the selected
264 features.
265
(...)
277 instance (e.g., GroupKFold).
278 """
--> 279 return self._fit(X, y, groups)
File C:\Anaconda3\envs\py38_skl\lib\site-packages\genetic_selection\gscv.py:343, in GeneticSelectionCV._fit(self, X, y, groups)
340 print("Selecting features with genetic algorithm.")
342 with np.printoptions(precision=6, suppress=True, sign=" "):
--> 343 _, log = _eaFunction(pop, toolbox, cxpb=self.crossover_proba,
344 mutpb=self.mutation_proba, ngen=self.n_generations,
345 ngen_no_change=self.n_gen_no_change,
346 stats=stats, halloffame=hof, verbose=self.verbose)
347 if self.n_jobs != 1:
348 pool.close()
File C:\Anaconda3\envs\py38_skl\lib\site-packages\genetic_selection\gscv.py:50, in _eaFunction(population, toolbox, cxpb, mutpb, ngen, ngen_no_change, stats, halloffame, verbose)
48 fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
49 for ind, fit in zip(invalid_ind, fitnesses):
---> 50 ind.fitness.values = fit
52 if halloffame is None:
53 raise ValueError("The 'halloffame' parameter should not be None.")
File C:\Anaconda3\envs\py38_skl\lib\site-packages\deap\base.py:188, in Fitness.setValues(self, values)
187 def setValues(self, values):
--> 188 assert len(values) == len(self.weights), "Assigned values have not the same length than fitness weights"
189 try:
190 self.wvalues = tuple(map(mul, values, self.weights))
AssertionError: Assigned values have not the same length than fitness weights
However, when I exclude dim__fs_wrapper from the pipeline, the error does not occur at all. The purpose of this transformer is to select a feature selection method from amongst 'passthrough' and estimators wrapped in SelectFromModel, RFECV and GeneticSelectionCV.