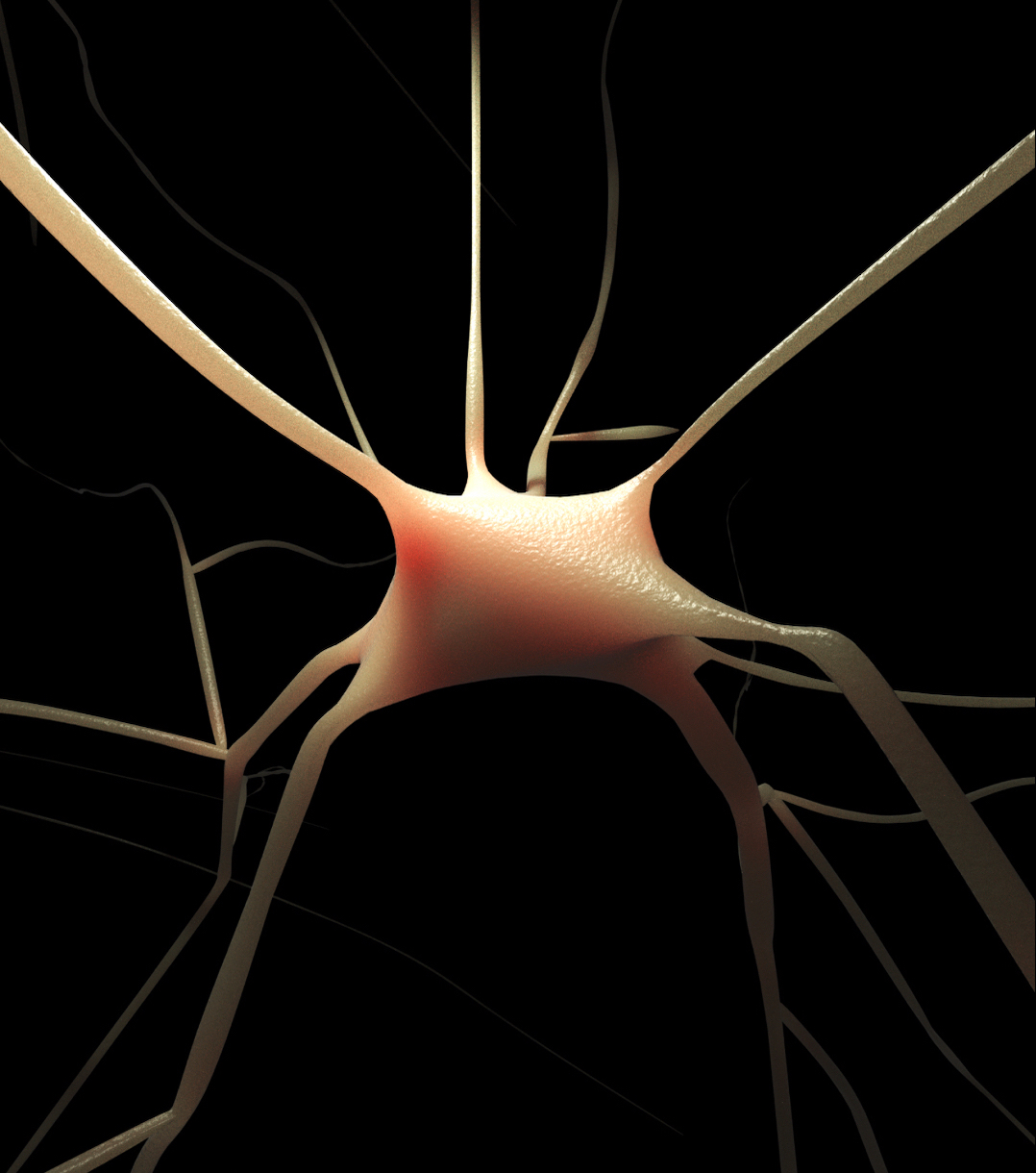
- Automated analysis of neuronal morphology skeletons that are digitally reconstructed from optical microscopy stacks.
- An easy context to load broken morphology skeletons and repair them manually.
- Sketching and building three-dimensional representations of the morphology skeletons using various methods for visual analytics.
- Dendrogram visualization and rendering.
- Visualization of spines and synapses for neuronal morphologies of digitally reconstructed BBP circuits using BluePy.
- Automated reconstruction of accurate three-dimensional somata profiles, even with classical morphology skeletons that do not have any three-dimensional data of their somata. This approach uses the physics engine of Blender based on Hooke's law and mass spring models.
- Automated reconstruction of polygonal mesh models that represent the membranes of the neuronal morphologies based on the piecewise meshing method presented by Abdellah et al., 2017.
- Accurate mesh reconstruction with metaballs to handle morphologies with irregular and small branching angles.
- Watertight mesh reconstruction using voxelization based remeshing (in Blender 3.0 onwards) for reaction-diffusion simulations.
- Automated generation of high quality media for scientific documents and publications using different shading styles and materials.
- High fidelity mesh reconstruction based on Skin modifiers as presented by Abdellah et al., 2019.
- Robust mesh reconstruction of synthetic astroglial cells using Metaballs and implicit geometries Abdellah et al., 2021
- Fast mesh reconstruction of spiny neuronal morohologies based on union operators for rendering transparent meshes Abdellah et al., 2022.
- Multiple interfaces: user-friendly graphical user interface, a rich command line interface, editable configuration files and a high level python API for python scripting.
- Importing morphologies in multiple file formats including SWC, H5, Neurolucida ASCII format or even from a BBP circuit using GIDs and cell targets.
- Exporting the reconstructed meshes in several file formats including PLY, OBJ, STL and also as a Blender file (.blend).
- Parallel batch processing on multi-node visualization clusters using SLURM workload manager.
- 06.2023: New logo release.
- 05.2023: Release 1.9.0
- 05.2023: Blender 2.79 and 2.8 are no longer supported by NeuroMorphoVis.
- 04.2023: Implementation of the Voxelization-based remeshing to create watertight mesh models of neurons and astrocytes.
- 03.2023: Adding the Synaptics panel to NeuroMorphoVis.
- 09.2022: Paper, Generating high fidelity surface meshes of neocortical neurons using skin modifiers, in EG Computer Graphics and Visual Computing (CGVC) 2022, more information.
- 04.2020: Release 1.4.5
- 09.2019: Paper: Generating High Fidelity Surface Meshes of Neocortical Neurons using Skin Modifiers, in EG Computer Graphics and Visual Computing (CGVC) 2019, more information.
- 05.2019: Release 1.3.1
- 03.2019: Release 1.3.0
- 10.2018: Release 1.0.0
- 07.2018: NeuroMorphoVis' core paper is published in Oxford Bioinformatics, more information.
NeuroMorphoVis is mainly based on Blender. Blender is a free software and can be downloaded from Blender.org. Blender is released under the GNU General Public License (GPL, or “free software”). The current version of NeuroMorphoVis is compatible with the following Blender versions:
Nevertheless, it is recommended to avoid version 2.79 and use version 3.5.
NeuroMorphoVis can be downloaded as a binary archive bundled within Blender that can be easily extracted and used out-of-the-box. The optional dependencies are already shiped within this archive using pip on each respective platform. This package (released every minor version update of the software) is recommended for Windows users or those who cannot use the Terminal. Otherwise, users can just download an installation script that will automatically install the entire package to a user-specified directory. This script does not require sudo permissions.
NeuroMorphoVis can be installed to a user-specified directory from a Unix (Linux or macOSX) terminal. Installation procedures are available in this page.
NeuroMorphoVis packages are available for Ubuntu, RedHat, macOSX and Windows from the releases page. After downloading the package, users can load NeuroMorphoVis into Blender as explained in this page.
NeuroMorphoVis is primarily designed as a plug-in in Blender. It comes with a user-friendly GUI and a rich set of command line options. Moreover, the tool is configurable via input configuration files making it possible to link it to web interface or using it on massively parallel visualization clusters for batch production.
The tool is also extensible via its powerful python API that can be imported in Blender console and text browser.
To make it accessible to end users with minimal programming knowledge or even with no programming experience at all, the core functionality of NeuroMorphoVis is exposed to users via a friendly graphical user interface that would allow them to navigate and adjust the parameters of the different toolboxes seamlessly. A detailed guide to use NeuroMorphoVis from its GUI is available in this user guide.
A screen shot of a neuronal morphology skeleton reconstructed by NeuroMorphoVis and sketched in the 3D view port of Blender. Note that the neurites are tagged with different colors (green for apical dendrite, red for basal dendrites and blue for the axon) and the soma (in yellow) is reconstructed as a realistic three-dimensional profile not as a sphere.NeuroMorphoVis has a rich command line interface that would make it easy to connect it to a web interface or use it to accomplish multiple tasks in an automated way. A list of all the command line options and their description are available in this user guide.
Users can easily configure and use NeuroMorphoVis via editable configuration files. Instructions to write and use configurations files are available in this user guide.
Please refer to the github issue tracker for fixed and open bugs. User can also report any bugs and request new features needed for their research. We are happy to provide direct support .
If you use NeuroMorphoVis for your research, media design or other purposes, please cite our paper NeuroMorphoVis: a collaborative framework for analysis and visualization of neuronal morphology skeletons reconstructed from microscopy stacks using the following entry:
@article{abdellah2018neuromorphovis,
title={NeuroMorphoVis: a collaborative framework for analysis and visualization of neuronal morphology
skeletons reconstructed from microscopy stacks},
author={Abdellah, Marwan and Hernando, Juan and Eilemann, Stefan and Lapere, Samuel and Antille,
Nicolas and Markram, Henry and Sch{\"u}rmann, Felix},
journal={Bioinformatics},
volume={34},
number={13},
pages={i574--i582},
year={2018},
publisher={Oxford University Press}
}
The supplementary material of this paper is also available in this document.
The core algorithms of the soma and mesh reconstruction modules are described in this paper Reconstruction and visualization of large-scale volumetric models of neocortical circuits for physically-plausible in silico optical studies
@article{abdellah2017reconstruction,
title={Reconstruction and visualization of large-scale volumetric models of neocortical circuits
for physically-plausible in silico optical studies},
author={Abdellah, Marwan and Hernando, Juan and Antille, Nicolas and Eilemann, Stefan and
Markram, Henry and Sch{\"u}rmann, Felix},
journal={BMC bioinformatics},
volume={18},
number={10},
pages={402},
year={2017},
publisher={BioMed Central}
}
The mesh generation algorithm with skin modifiers is described in this paper Generating High Fidelity Surface Meshes of Neocortical Neurons using Skin Modifiers
@inproceedings{abdellah2019generating,
booktitle={Computer Graphics and Visual Computing (CGVC)},
editor={Vidal, Franck P. and Tam, Gary K. L. and Roberts, Jonathan C.},
title={Generating High Fidelity Surface Meshes of Neocortical Neurons using Skin Modifiers},
author={Abdellah, Marwan and Favreau, Cyrille and Hernando, Juan and Lapere, Samuel and Schürmann, Felix},
year={2019},
publisher={The Eurographics Association},
isbn={978-3-03868-096-3},
doi={10.2312/cgvc.20191257}
}
The applications of NeuroMorhoVis are deomnstrated in the thesis In silico Brain Imaging: Physically-plausible Methods for Visualizing Neocortical Microcircuitry
@phdthesis{abdellah2017insilico:232444,
title = {In Silico Brain Imaging Physically-plausible Methods for Visualizing
Neocortical Microcircuitry},
author = {Abdellah, Marwan},
publisher = {EPFL},
address = {Lausanne},
pages = {400},
year = {2017}
}
An exhaustive list of all the publications that use or mention NeuroMorphoVis is available in this Wiki page.
NeuroMorphoVis is developed by the Visualization team at the Blue Brain Project, Ecole Polytechnique Federale de Lausanne (EPFL) as part of Marwan Abdellah's PhD (In silico Brain Imaging: Physically-plausible Methods for Visualizing Neocortical Microcircuitry). Financial support was provided by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology, and by competitive research funding from King Abdullah University of Science and Technology (KAUST).
NeuroMorphoVis is available to download and use under the GNU General Public License (GPL, or “free software”). The code is open sourced with approval from the open sourcing committee and principal coordinators of the Blue Brain Project in December 2019.
Copyright (c) 2016-2023 Blue Brain Project/EPFL
-
Blender (C) is copyright to Blender Foundation. The Blender Foundation is a non-profit organization responsible for the development of Blender. Blender is released under the GNU Public License, as Free Software, and therefore can be distributed by anyone freely.
-
Slurm, is a free and open-source job scheduler for Linux and Unix-like kernels, used by many of the world's supercomputers and computer clusters. Slurm is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation.
-
The SWC morphology samples are available from NeuroMorpho.Org. NeuroMorpho.Org is a centrally curated inventory or repository of digitally reconstructed neurons associated with peer-reviewed publications.
-
The H5 morphology samples are available with permissions from the Blue Brain Project, Ecole Polytechnique Federale de Lausanne (EPFL).
-
The analysis plots are created based on Matplotlib and Seaborn. Matplotlib is a comprehensive library for creating static, animated, and interactive visualizations in Python and Seaborn is a Python data visualization library based on matplotlib. It provides a high-level interface for drawing attractive and informative statistical graphics. Matplotlib only uses BSD compatible code, and its license is based on the PSF license. Seaborn uses BSD license.
-
The analysis distributions are gathered into a single PDF using PyPDF2. PyPDF2 is a pure-python PDF library capable of splitting, merging together, cropping, and transforming the pages of PDF files. PyPDF2 uses the BSD License.
-
The table of contents for all the user documentation pages are generated with markdown-toc.
For more information on NeuroMorphoVis, comments or suggestions, please contact:
Marwan Abdellah
Scientific Visualiation Expert
Visualization Team, Computing Division
Blue Brain Project
[email protected]
marwan-abdellah.com
-
Starting
-
Panels
-
Other Links