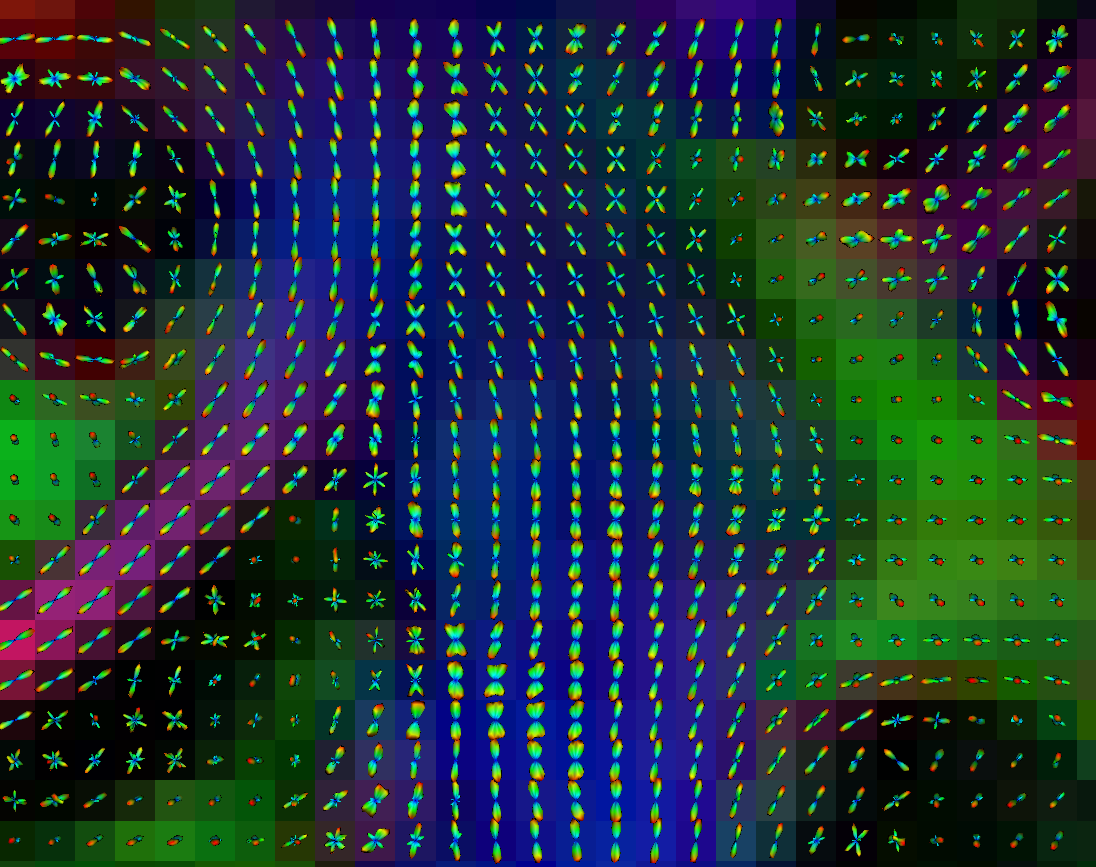
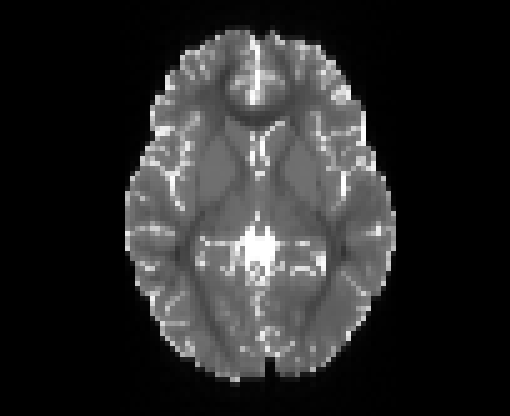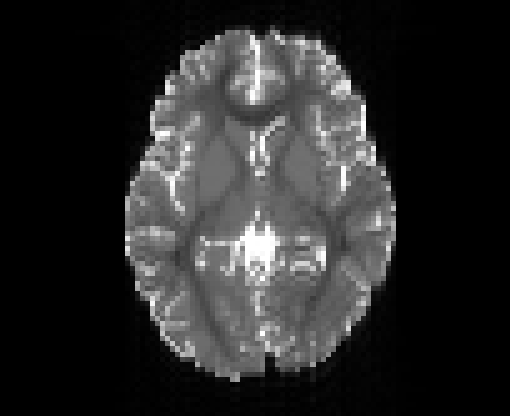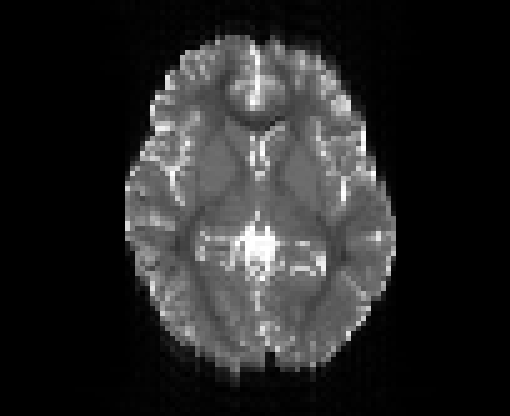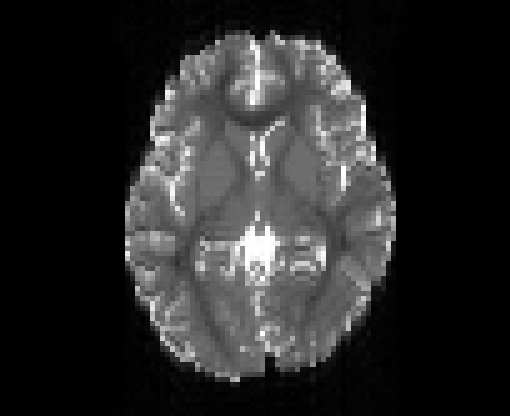
Copyright © German Cancer Research Center (DKFZ), Division of Medical Image Computing (MIC). Please make sure that your usage of this code is in compliance with the code license.
The MITK Diffusion application [1,2] offers a selection of image analysis algorithms for the processing of diffusion-weighted MR images. It encompasses the research of the Division of Medical Image Computing at the German Cancer Research Center (DKFZ).
- Downloads
- Requirements
- Features
- Related Links
- Image Gallery
- Building MITK Diffusion from source
- User Manual
- Report a Bug
- References
- Contact
Please have a look at the requirements for running MITK Diffusion with all its features successfully!
The latest builds come as executable setup wizards that install MITK Diffusion on your system or alternatively as simple .tar.gz or .zip archive where you can execute MITK Diffusion and the command line apps "manually". You can find the newest installers here.
If you encounter any bugs, please report them here on github or use the MITK-users mailing list. We are grateful for any feedback!
For Windows users: MITK Diffusion requires the Microsoft Visual C++ 2017 Redistributable to be installed on the system. The MITK Diffusion installer automatically installs this redistributable for you if not already present on the system, but it needs administrative privileges to do so. So to install the redistributable, run the MITK Diffusion installer as administrator.
Support for most established image formats
- Images: DICOM, NIFTI, NRRD (peak and SH images compatible with MRtrix)
- Tractograms: fib/vtk, tck and trk.
Image preprocessing
- Registration
- Head-motion correction
- Denoising
- Resampling, cropping, flipping and merging
- Header modifications
- Single volume extraction
Diffusion gradient/b-value processing
- b-value rounding
- Gradient direction flipping
- Gradient direction subsampling
- Averaging of gradient directions/volumes
- Gradient direction and b-value visualization
ODF reconstruction and signal modelling
- Tensor and Q-ball reconstruction
- ODF peak calculation
- MRtrix or camino results can be imported
Quantification of diffusion-weighted/tensor/ODF images
- Intravoxel Incoherent Motion (IVIM) and diffusion kurtosis analysis
- Calculation of many other derived indices such as ADC, MD, GFA, FA, RA, AD, RD
- Image statistics
Segmentation
- Manual image segmentation and operations on segmentations
Fiber tractography
- Global tractography [4]
- Streamline tractography
- Interactive (similar to [5]) or seed image based
- Deterministic or probabilistic
- Peak, ODF, tensor and raw dMRI based. The latter one in conjunction with machine learning based tractography [6]
- Various possibilities for anatomical constraints.
- Tractography priors in form of additional peak images, e.g. obtained using TractSeg
Fiber processing
- Tract dissection (parcellation or ROI based)
- Tract filtering by
- length
- curvature
- direction
- weight
- density
- Tract resampling and compression
- Tract transformation
- Mirroring
- Rotating and translating
- Registration (apply transform of previously performed image registration)
- Tract coloring
- Curvature
- Length
- Weight
- Scalar map (e.g. FA)
- Other operations
- Join
- Subtract
- Copy
- Fiber clustering [7]
- Fiber fitting and weighting similar to SIFT2 and LiFE [8,9]
- Principal direction extraction (fibers --> peaks)
- Tract derived images:
- Tract density images
- Tract endpoint images
- Tract envelopes
Fiberfox dMRI simulations [10]
- Multi-compartment signal modeling
- Simulation of the k-space acquisition including
- Compartment specific relaxation effects
- Artifacts such as noise, spikes, ghosts, aliasing, distortions, signal drift, head motion, eddy currents and Gibbs ringing
- Definition of important acquisition parameters such as bvalues and gradient directions, TE, TR, dwell time, partial Fourier, ...
- Manual definition of fiber configurations, e.g. for evaluation purposes
- Automatic generation of random fiber configurations
Other features
- Integrated screenshot maker
- Command line tools for most functionalities
- Great python package for logging your (MITK or other) command line experiments:
- https://github.com/MIC-DKFZ/cmdint
pip3 install cmdint
- TractSeg reference data of 72 semiautomatically defined bundles in 105 HCP subjects: https://zenodo.org/record/1285152
- TractSeg python package: https://github.com/MIC-DKFZ/TractSeg
- Simulated dMRI images and ground truth of random fiber phantoms in various configurations: https://doi.org/10.5281/zenodo.2533250
- ISMRM 2015 Tractography Challenge Data: https://doi.org/10.5281/zenodo.572345 & https://doi.org/10.5281/zenodo.1007149
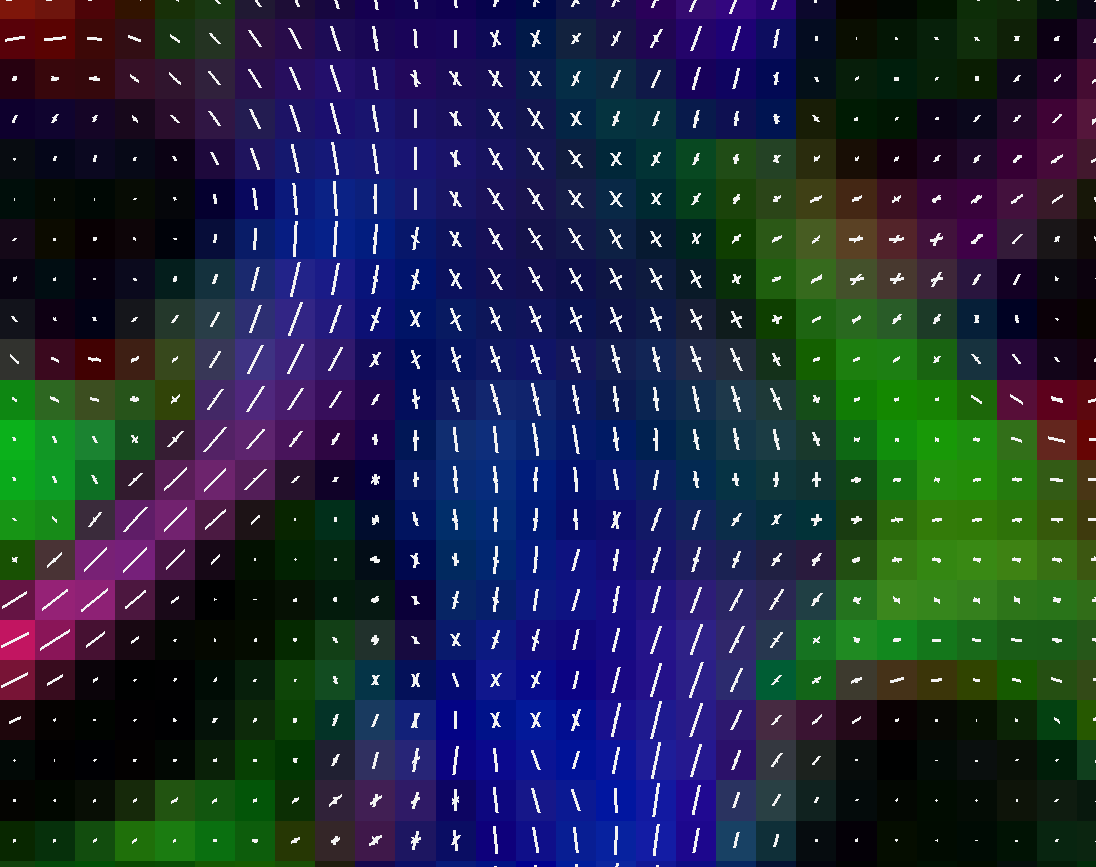
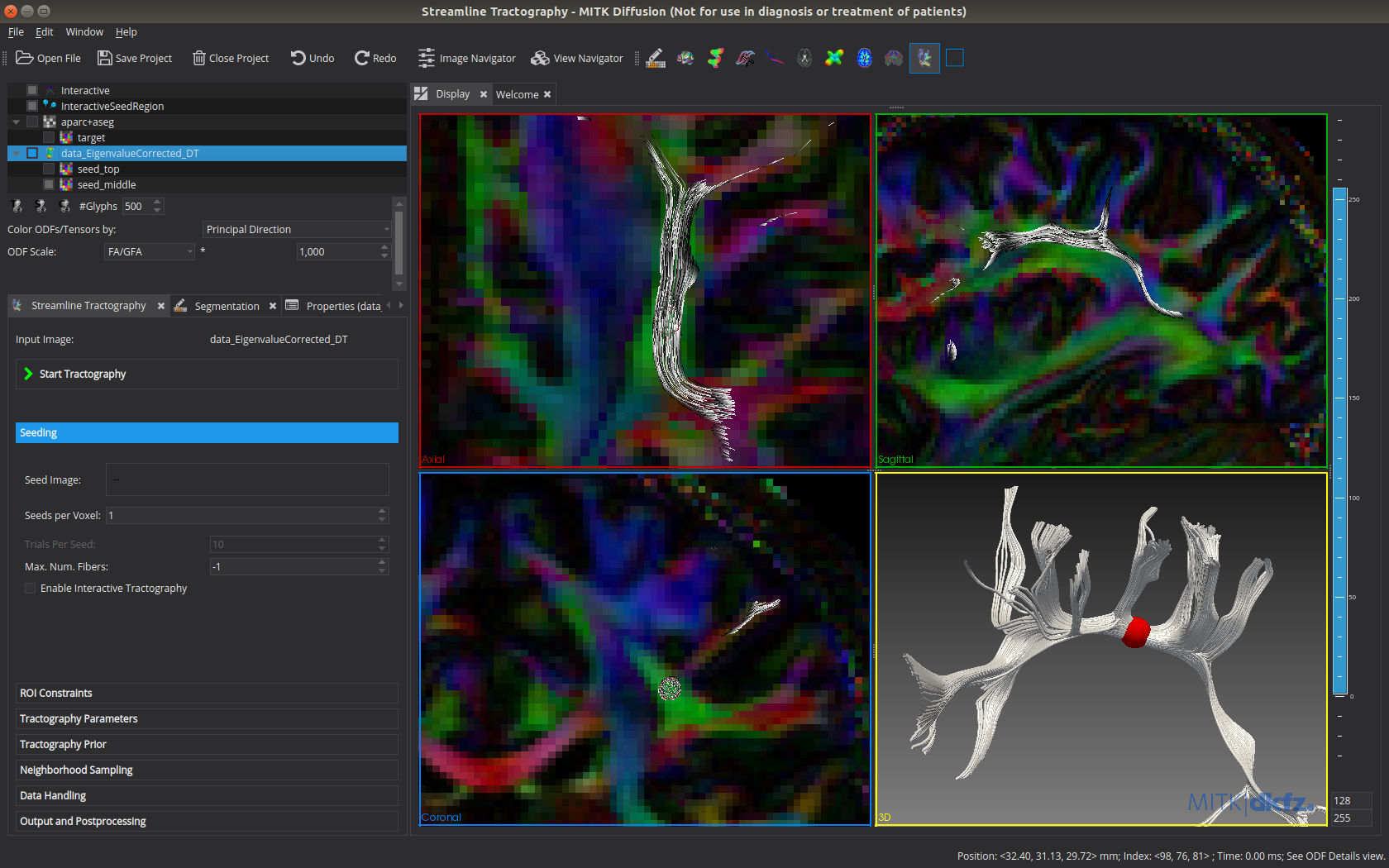
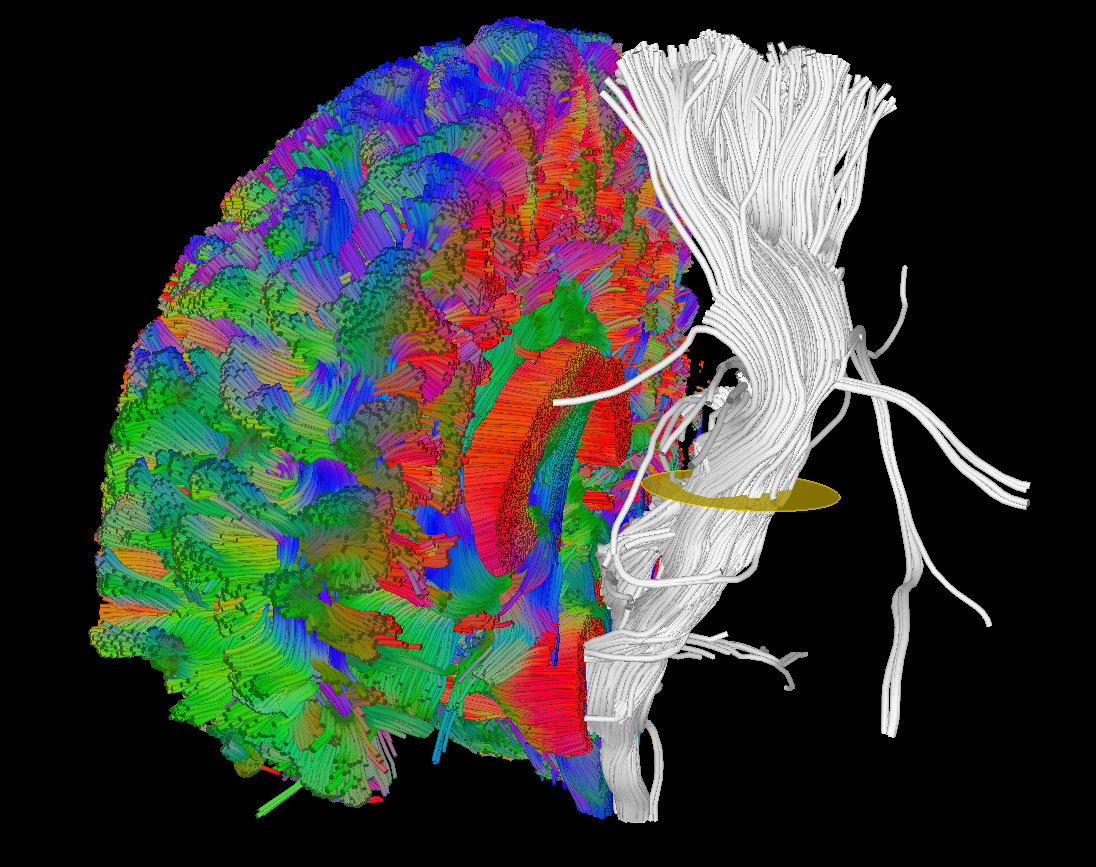


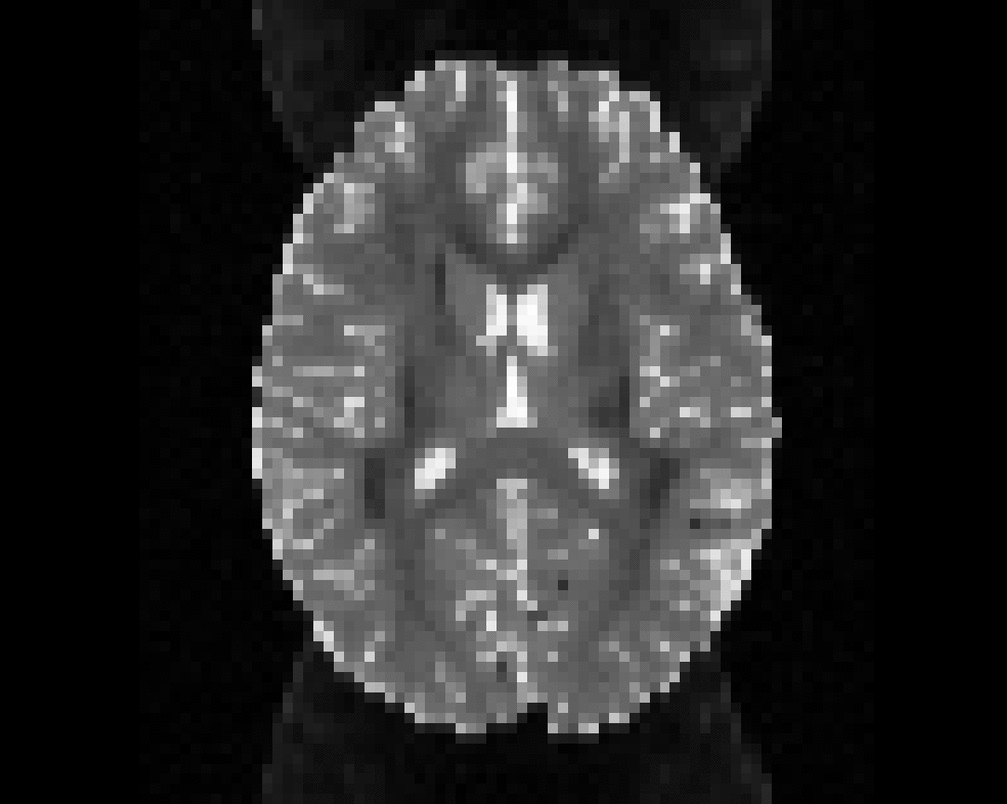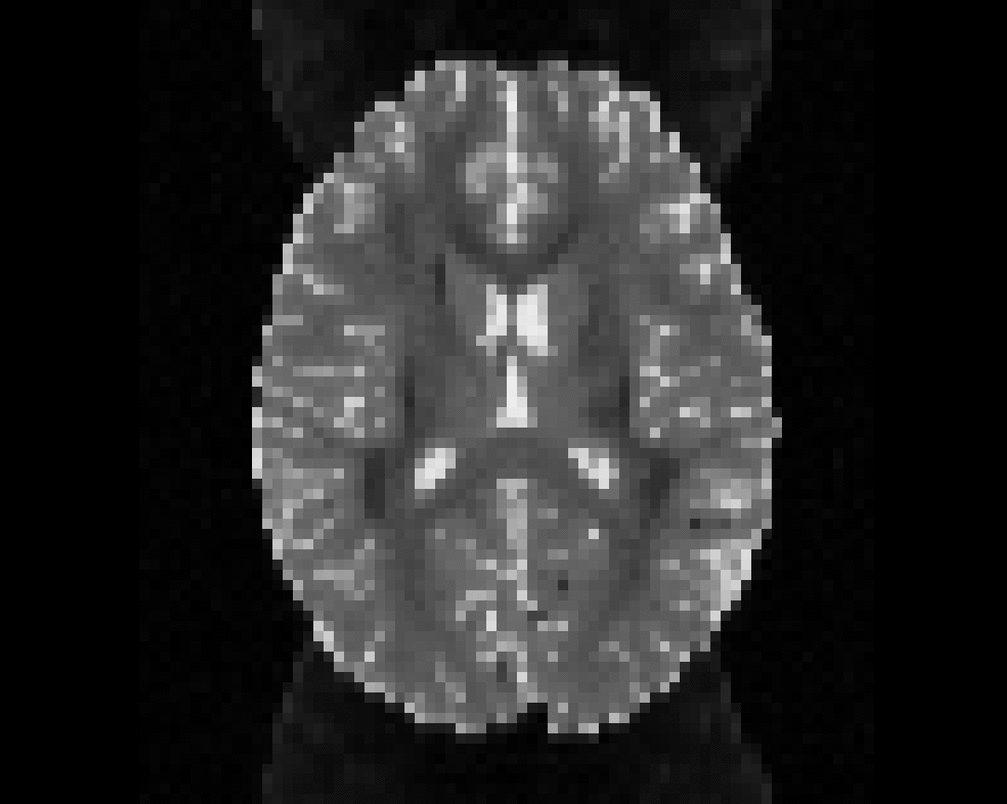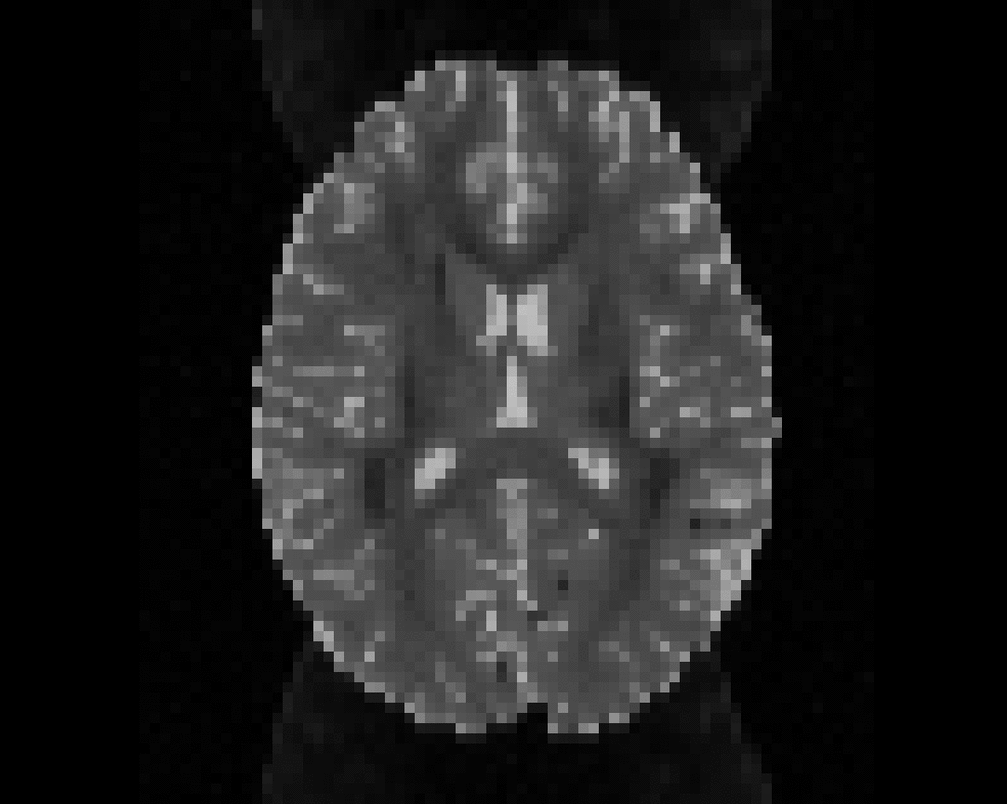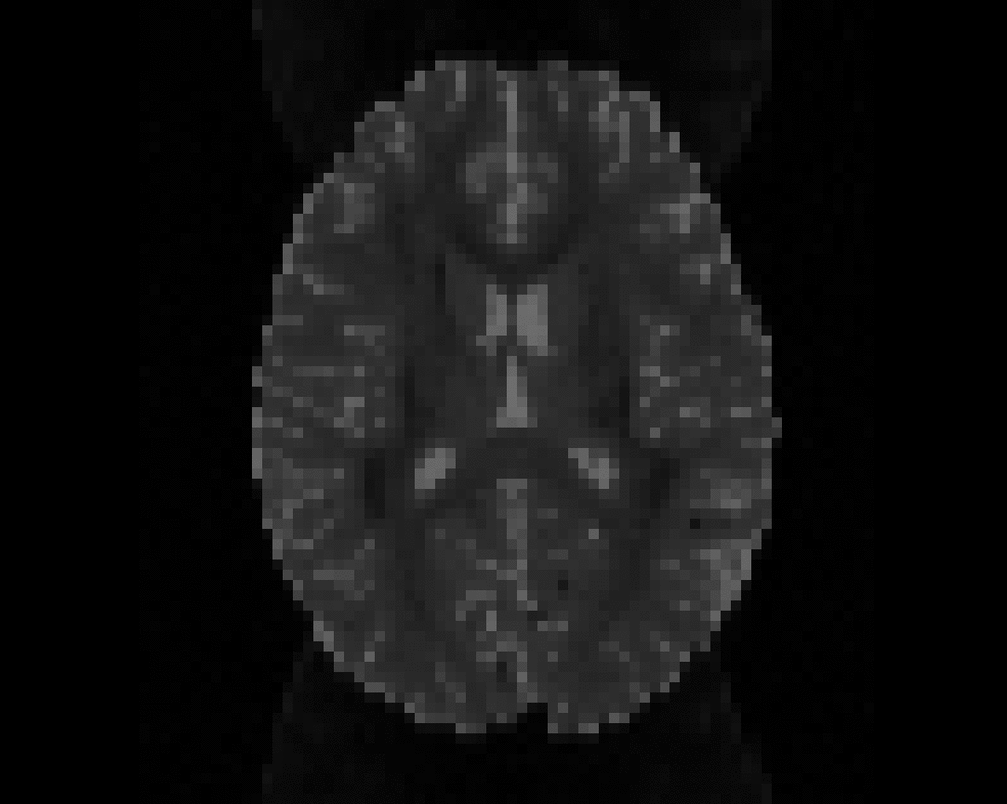
Illustration of simulated dMRI images with various artifacts (a bit excessive for illustration purposes): eddy current distortions (1), motion and spike (2), intensity drift (3), motion, eddy and noise (4), ringing (5), B0 inhomogeneity distortions (6), from left to right.

Please not that we moved away from a separate MITK Diffusion application and are instead building configurations of the MITK Workbench. In a (near)-future version of MITK, customizations of the Workbench will be possible to change the display and executable name etc. A workaround is to use the MITK branch https://github.com/MITK/MITK/tree/feature/T30337-WorkbenchCustomizations2, which already enables some of those customizations but the final mechanism fot that will be different.
-
Install Qt on your system (currently 6.6.1).
-
Clone MITK from github using Git version control.
-
Clone MITK Diffusion from github.
-
Configure the MITK Superbuild using CMake.
- Choose the MITK source code directory and an empty binary directory.
- Click "Configure".
- Set the option MITK_EXTENSION_DIRS to "/path/to/my/mitk-diffusion-repository".
- Click "Configure".
- Set the option MITK_BUILD_CONFIGURATION to "DiffusionRelease" or any other desired configuration. There are various configurations to build e.g. only IVIM modules or only tractography. The DiffusionRelease/DiffusionDebug configurations contain all modules.
- Click "Generate".
-
Start the Superbuild:
- Linux: Open a console window, navigate to the build folder and type "make -j8" (optionally supply the number threads to be used for a parallel build with -j).
- Windows (requires visual studio): Open the MITK Superbuild solution file and build all projects.
- The Superbuild may take some time.
- After the Superbuild has finished, change the cmake binary directory from "/path/to/my/build" to "/path/to/my/build/MITK-build" and configure+generate again.
- Build again in "/path/to/my/build/MITK-build" using make (linux/mac) or the VS solution file.
- The build may again take some time and should yield the binaries in "/path/to/my/build/MITK-build/bin"
More detailed build instructions can be found in the documentation.
Continuous integration: https://cdash.mitk.org/index.php?project=MITK-Diffusion
All publications of the Division of Medical Image Computing can be found [https://www.dkfz.de/en/mic/publications/ here].
[1] Fritzsche, Klaus H., Peter F. Neher, Ignaz Reicht, Thomas van Bruggen, Caspar Goch, Marco Reisert, Marco Nolden, et al. “MITK Diffusion Imaging.” Methods of Information in Medicine 51, no. 5 (2012): 441.
[2] Fritzsche, K., and H.-P. Meinzer. “MITK-DI A New Diffusion Imaging Component for MITK.” In Bildverarbeitung Für Die Medizin, n.d.
[3] Wasserthal, Jakob, Peter Neher, and Klaus H. Maier-Hein. “TractSeg - Fast and Accurate White Matter Tract Segmentation.” NeuroImage 183 (August 4, 2018): 239–53.
[4] Neher, P. F., B. Stieltjes, M. Reisert, I. Reicht, H.P. Meinzer, and K. Maier-Hein. “MITK Global Tractography.” In SPIE Medical Imaging: Image Processing, 2012.
[5] Chamberland, M., K. Whittingstall, D. Fortin, D. Mathieu, und M. Descoteaux. „Real-time multi-peak tractography for instantaneous connectivity display“. Front Neuroinform 8 (2014): 59. doi:10.3389/fninf.2014.00059.
[6] Neher, Peter F., Marc-Alexandre Côté, Jean-Christophe Houde, Maxime Descoteaux, and Klaus H. Maier-Hein. “Fiber Tractography Using Machine Learning.” NeuroImage. Accessed July 17, 2017. doi:10.1016/j.neuroimage.2017.07.028.
[7] Garyfallidis, Eleftherios, Matthew Brett, Marta Morgado Correia, Guy B. Williams, and Ian Nimmo-Smith. “QuickBundles, a Method for Tractography Simplification.” Frontiers in Neuroscience 6 (2012).
[8] Smith, Robert E., Jacques-Donald Tournier, Fernando Calamante, and Alan Connelly. “SIFT2: Enabling Dense Quantitative Assessment of Brain White Matter Connectivity Using Streamlines Tractography.” NeuroImage 119, no. Supplement C (October 1, 2015): 338–51.
[9] Pestilli, Franco, Jason D. Yeatman, Ariel Rokem, Kendrick N. Kay, and Brian A. Wandell. “Evaluation and Statistical Inference for Human Connectomes.” Nature Methods 11, no. 10 (October 2014): 1058–63.
[10] Neher, Peter F., Frederik B. Laun, Bram Stieltjes, and Klaus H. Maier-Hein. “Fiberfox: Facilitating the Creation of Realistic White Matter Software Phantoms.” Magnetic Resonance in Medicine 72, no. 5 (November 2014): 1460–70. doi:10.1002/mrm.25045.
If you have questions about the application or if you would like to give us feedback, don't hesitate to contact us using our mailing list or, for questions that are of no interest for the community, directly.