EEG-DL is a Deep Learning (DL) library written by TensorFlow for EEG Tasks (Signals) Classification. It provides the latest DL algorithms and keeps updated.
- Documentation
- Usage Demo
- Notice
- Research Ideas
- Common Issues
- Structure of the Code
- Citation
- Other Useful Resources
- Contribution
- Organizations
The supported models include
| No. | Model | Codes |
|---|---|---|
| 1 | Deep Neural Networks | DNN |
| 2 | Convolutional Neural Networks [Paper] [Tutorial] | CNN |
| 3 | Deep Residual Convolutional Neural Networks [Paper] | ResNet |
| 4 | Thin Residual Convolutional Neural Networks [Paper] | Thin ResNet |
| 5 | Densely Connected Convolutional Neural Networks [Paper] | DenseNet |
| 6 | Fully Convolutional Neural Networks [Paper] | FCN |
| 7 | One Shot Learning with Siamese Networks (CNNs Backbone) [Paper] [Tutorial] |
Siamese Networks |
| 8 | Graph Convolutional Neural Networks [Paper] [Presentation] |
GCN / Graph CNN |
| 9 | Deep Residual Graph Convolutional Neural Networks [Paper] | ResGCN |
| 10 | Densely Connected Graph Convolutional Neural Networks | DenseGCN |
| 11 | Bayesian Convolutional Neural Network via Variational Inference [Paper] |
Bayesian CNNs |
| 12 | Recurrent Neural Networks [Paper] | RNN |
| 13 | Attention-based Recurrent Neural Networks [Paper] | RNN with Attention |
| 14 | Bidirectional Recurrent Neural Networks [Paper] | BiRNN |
| 15 | Attention-based Bidirectional Recurrent Neural Networks [Paper] | BiRNN with Attention |
| 16 | Long-short Term Memory [Paper] | LSTM |
| 17 | Attention-based Long-short Term Memory [Paper] | LSTM with Attention |
| 18 | Bidirectional Long-short Term Memory [Paper] | BiLSTM |
| 19 | Attention-based Bidirectional Long-short Term Memory [Paper] | BiLSTM with Attention |
| 20 | Gated Recurrent Unit [Paper] | GRU |
| 21 | Attention-based Gated Recurrent Unit [Paper] | GRU with Attention |
| 22 | Bidirectional Gated Recurrent Unit [Paper] | BiGRU |
| 23 | Attention-based Bidirectional Gated Recurrent Unit [Paper] | BiGRU with Attention |
| 24 | Attention-based BiLSTM + GCN [Paper] | Attention-based BiLSTM GCN |
| 25 | Transformer [Paper] [Paper] | Transformer |
| 26 | Transfer Learning with Transformer (This code is only for reference!) (You can modify the codes to fit your data.) |
Stage 1: Pre-training Stage 2: Fine Tuning |
One EEG Motor Imagery (MI) benchmark is currently supported. Other benchmarks in the field of EEG or BCI can be found here.
| No. | Dataset | Tutorial |
|---|---|---|
| 1 | EEG Motor Movement/Imagery Dataset | Tutorial |
The evaluation criteria consists of
| Evaluation Metrics | Tutorial |
|---|---|
| Confusion Matrix | Tutorial |
| Accuracy / Precision / Recall / F1 Score / Kappa Coefficient | Tutorial |
| Receiver Operating Characteristic (ROC) Curve / Area under the Curve (AUC) | - |
| Paired-wise t-test via R language | Tutorial |
The evaluation metrics are mainly supported for four-class classification. If you wish to switch to two-class or three-class classification, please modify this file to adapt to your personal Dataset classes. Meanwhile, the details about the evaluation metrics can be found in this paper.
-
(Under Any Python Environment) Download the EEG Motor Movement/Imagery Dataset via this script.
$ python MIND_Get_EDF.py -
(Under Python 2.7 Environment) Read the .edf files (One of the raw EEG signals formats) and save them into Matlab .m files via this script. FYI, this script must be executed under the Python 2 environment (Python 2.7 is recommended) due to some Python 2 syntax. If using Python 3 environment to run the file, there might be no error, but the labels of EEG tasks would be totally messed up.
$ python Extract-Raw-Data-Into-Matlab-Files.py -
Preprocessed the Dataset via the Matlab and save the data into the Excel files (training_set, training_label, test_set, and test_label) via these scripts with regards to different models. FYI, every lines of the Excel file is a sample, and the columns can be regarded as features, e.g., 4096 columns mean 64 channels X 64 time points. Later, the models will reshape 4096 columns into a Matrix with the shape 64 channels X 64 time points. You should can change the number of columns to fit your own needs, e.g., the real dimension of your own Dataset.
-
(Prerequsites) Train and test deep learning models under the Python 3.6 Environment (Highly Recommended) for EEG signals / tasks classification via the EEG-DL library, which provides multiple SOTA DL models.
Python Version: Python 3.6 (Recommended) TensorFlow Version: TensorFlow 1.13.1Use the below command to install TensorFlow GPU Version 1.13.1:
$ pip install --upgrade --force-reinstall tensorflow-gpu==1.13.1 --user
-
Read evaluation criterias (through iterations) via the Tensorboard. You can follow this tutorial. When you finished training the model, you will find the "events.out.tfevents.***" in the folder, e.g., "/Users/shuyuej/Desktop/trained_model/". You can use the following command in your terminal:
$ tensorboard --logdir="/Users/shuyuej/Desktop/trained_model/" --host=127.0.0.1
You can open the website in the Google Chrome (Highly Recommended).
http://127.0.0.1:6006/
Then you can read and save the criterias into Excel .csv files.
-
Finally, draw beautiful paper photograph using Matlab or Python. Please follow these scripts.
-
I have tested all the files (Python and Matlab) under the macOS. Be advised that for some Matlab files, several Matlab functions are different between Windows Operating System (OS) and macOS. For example, I used "readmatrix" function to read CSV files in the MacOS. However, I have to use “csvread” function in the Windows because there was no such "readmatrix" Matlab function in the Windows. If you have met similar problems, I recommend you to Google or Baidu them. You can definitely work them out.
-
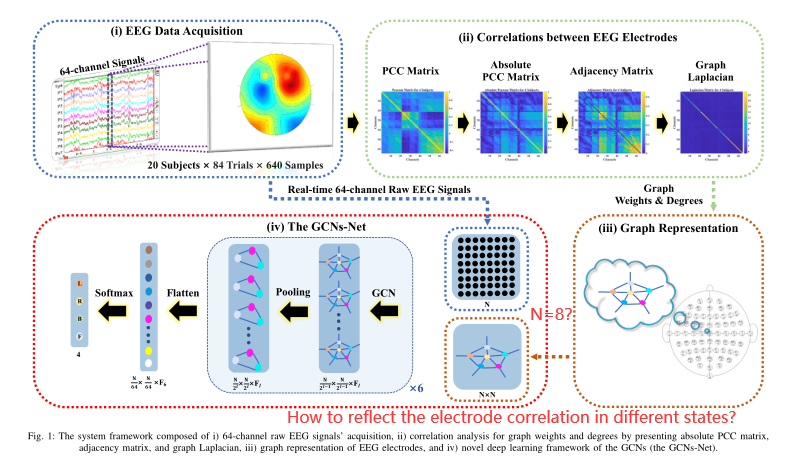
For the GCNs-Net (GCN Model), for the graph Convolutional layer, the dimensionality of the graph will be unchanged, and for the max-pooling layer, the dimensionality of the graph will be reduced by 2. That means, if you have N X N graph Laplacian, after the max-pooling layer, the dimension will be N/2 X N/2. If you have a 15-channel EEG system, it cannot use max-pooling unless you selected 14 --> 7 or 12 --> 6 --> 3 or 10 --> 5 or 8 --> 4 --> 2 --> 1, etc. The details can be reviewed from this paper.
-
The Loss Function can be changed or modified from this file.
-
The Dataset Loader can be changed or modified from this file.
-
Dynamic Graph Convolutional Neural Networks [Paper Survey] [Paper Reading]
-
Neural Architecture Search / AutoML (Automatic Machine Learning) [Tsinghua AutoGraph]
-
Reinforcement Learning Algorithms (e.g., Deep Q-Learning) [Tsinghua Tianshou] [Doc for Chinese Readers]
-
Bayesian Convolutional Neural Networks [Paper] [Thesis] [Codes]
-
Transformer / Self-attention / Non-local Modeling [Paper Collections] [Transformer Codes] [Non-local Modeling PyTorch Codes]
[Why Non-local Modeling?] [Paper] [A Detailed Presentation] [Slides] [Poster]
-
Self-supervised Learning + Transformer [Presentation]
-
ValueError: Cannot feed value of shape (1024, 1) for Tensor 'input/label:0', which has shape '(1024,)'
To solve this issue, you have to squeeze the shape of the labels from (1024, 1) to (1024,) using np.squeeze. Please edit the DataLoader.py file. From original codes:
train_labels = pd.read_csv(DIR + 'training_label.csv', header=None) train_labels = np.array(train_labels).astype('float32') test_labels = pd.read_csv(DIR + 'test_label.csv', header=None) test_labels = np.array(test_labels).astype('float32')
to
train_labels = pd.read_csv(DIR + 'training_label.csv', header=None) train_labels = np.array(train_labels).astype('float32') train_labels = np.squeeze(train_labels) test_labels = pd.read_csv(DIR + 'test_label.csv', header=None) test_labels = np.array(test_labels).astype('float32') test_labels = np.squeeze(test_labels)
-
InvalidArgumentError: Nan in summary histogram for training/logits/bias/gradients
To solve this issue, you have to comment all the histogram summary. Please edit the GCN_Model.py file.
# Comment the above tf.summary.histogram from the GCN_Model.py File # # Histograms. # for grad, var in grads: # if grad is None: # print('warning: {} has no gradient'.format(var.op.name)) # else: # tf.summary.histogram(var.op.name + '/gradients', grad) def _weight_variable(self, shape, regularization=True): initial = tf.truncated_normal_initializer(0, 0.1) var = tf.get_variable('weights', shape, tf.float32, initializer=initial) if regularization: self.regularizers.append(tf.nn.l2_loss(var)) # tf.summary.histogram(var.op.name, var) return var def _bias_variable(self, shape, regularization=True): initial = tf.constant_initializer(0.1) var = tf.get_variable('bias', shape, tf.float32, initializer=initial) if regularization: self.regularizers.append(tf.nn.l2_loss(var)) # tf.summary.histogram(var.op.name, var) return var
-
TypeError: len() of unsized object
To solve this issue, you have to change the coarsen level to your own needs, and you can definitely change it to see the difference. Please edit the main-GCN.py file. For example, if you want to implement the GCNs-Net to a 10-channel EEG system, you have to set "levels" equal to 1 or 0 because there is at most only one max-pooling (10 --> 5). And you can change argument "level" to 1 or 0 to see the difference.
# This is the coarsen levels, you can definitely change the level to observe the difference graphs, perm = coarsening.coarsen(Adjacency_Matrix, levels=5, self_connections=False)
to
# This is the coarsen levels, you can definitely change the level to observe the difference graphs, perm = coarsening.coarsen(Adjacency_Matrix, levels=1, self_connections=False)
-
tensorflow.python.framework.errors_impl.InvalidArgumentError: Received a label value of 7 which is outside the valid range of [0, 7). Label values: 5 2 3 3 1 5 5 4 7 4 2 2 1 7 5 6 3 4 2 4
To solve this issue, for the GCNs-Net, when you make your dataset, you have to make your labels from 0 rather than 1. For example, if you have seven classes, your labels should be 0 (First class), 1 (Second class), 2 (Third class), 3 (Fourth class), 4 (Fifth class), 5 (Sixth class), 6 (Seventh class) instead of 1, 2, 3, 4, 5, 6, 7.
-
IndexError: list index out of range
To solve this issue, first of all, please double-check your Python Environment. Python 2.7 Environment is required. Besides, please install 0.1.11 version of pyEDFlib. The installation instruction is as follows:
$ pip install pyEDFlib==0.1.11
At the root of the project, you will see:
├── Download_Raw_EEG_Data
│ ├── Extract-Raw-Data-Into-Matlab-Files.py
│ ├── MIND_Get_EDF.py
│ ├── README.md
│ └── electrode_positions.txt
├── Draw_Photos
│ ├── Draw_Accuracy_Photo.m
│ ├── Draw_Box_Photo.m
│ ├── Draw_Confusion_Matrix.py
│ ├── Draw_Loss_Photo.m
│ ├── Draw_ROC_and_AUC.py
│ └── figure_boxplot.m
├── LICENSE
├── Logo.png
├── MANIFEST.in
├── Models
│ ├── DatasetAPI
│ │ └── DataLoader.py
│ ├── Evaluation_Metrics
│ │ └── Metrics.py
│ ├── Initialize_Variables
│ │ └── Initialize.py
│ ├── Loss_Function
│ │ └── Loss.py
│ ├── Network
│ │ ├── BiGRU.py
│ │ ├── BiGRU_with_Attention.py
│ │ ├── BiLSTM.py
│ │ ├── BiLSTM_with_Attention.py
│ │ ├── BiRNN.py
│ │ ├── BiRNN_with_Attention.py
│ │ ├── CNN.py
│ │ ├── DNN.py
│ │ ├── DenseCNN.py
│ │ ├── Fully_Conv_CNN.py
│ │ ├── GRU.py
│ │ ├── GRU_with_Attention.py
│ │ ├── LSTM.py
│ │ ├── LSTM_with_Attention.py
│ │ ├── RNN.py
│ │ ├── RNN_with_Attention.py
│ │ ├── ResCNN.py
│ │ ├── Siamese_Network.py
│ │ ├── Thin_ResNet.py
│ │ └── lib_for_GCN
│ │ ├── DenseGCN_Model.py
│ │ ├── GCN_Model.py
│ │ ├── ResGCN_Model.py
│ │ ├── coarsening.py
│ │ └── graph.py
│ ├── __init__.py
│ ├── main-BiGRU-with-Attention.py
│ ├── main-BiGRU.py
│ ├── main-BiLSTM-with-Attention.py
│ ├── main-BiLSTM.py
│ ├── main-BiRNN-with-Attention.py
│ ├── main-BiRNN.py
│ ├── main-CNN.py
│ ├── main-DNN.py
│ ├── main-DenseCNN.py
│ ├── main-DenseGCN.py
│ ├── main-FullyConvCNN.py
│ ├── main-GCN.py
│ ├── main-GRU-with-Attention.py
│ ├── main-GRU.py
│ ├── main-LSTM-with-Attention.py
│ ├── main-LSTM.py
│ ├── main-RNN-with-Attention.py
│ ├── main-RNN.py
│ ├── main-ResCNN.py
│ ├── main-ResGCN.py
│ ├── main-Siamese-Network.py
│ └── main-Thin-ResNet.py
├── NEEPU.png
├── Preprocess_EEG_Data
│ ├── For-CNN-based-Models
│ │ └── make_dataset.m
│ ├── For-DNN-based-Models
│ │ └── make_dataset.m
│ ├── For-GCN-based-Models
│ │ └── make_dataset.m
│ ├── For-RNN-based-Models
│ │ └── make_dataset.m
│ └── For-Siamese-Network-One-Shot-Learning
│ └── make_dataset.m
├── README.md
├── Saved_Files
│ └── README.md
├── requirements.txt
└── setup.py
If you find our library useful, please considering citing our papers in your publications. We provide a BibTeX entry below.
@article{hou2022gcn,
title = {{GCNs-Net}: A Graph Convolutional Neural Network Approach for Decoding Time-Resolved EEG Motor Imagery Signals},
author = {Hou, Yimin and Jia, Shuyue and Lun, Xiangmin and Hao, Ziqian and Shi, Yan and Li, Yang and Zeng, Rui and Lv, Jinglei},
journal = {IEEE Transactions on Neural Networks and Learning Systems},
volume = {},
number = {},
pages = {1-12},
year = {Sept. 2022},
doi = {10.1109/TNNLS.2022.3202569}
}
@article{hou2020novel,
title = {A Novel Approach of Decoding EEG Four-class Motor Imagery Tasks via Scout {ESI} and {CNN}},
author = {Hou, Yimin and Zhou, Lu and Jia, Shuyue and Lun, Xiangmin},
journal = {Journal of Neural Engineering},
volume = {17},
number = {1},
pages = {016048},
year = {Feb. 2020},
publisher = {IOP Publishing},
doi = {10.1088/1741-2552/ab4af6}
}
@article{hou2022deep,
title = {Deep Feature Mining via the Attention-Based Bidirectional Long Short Term Memory Graph Convolutional Neural Network for Human Motor Imagery Recognition},
author = {Hou, Yimin and Jia, Shuyue and Lun, Xiangmin and Zhang, Shu and Chen, Tao and Wang, Fang and Lv, Jinglei},
journal = {Frontiers in Bioengineering and Biotechnology},
volume = {9},
year = {Feb. 2022},
url = {https://www.frontiersin.org/article/10.3389/fbioe.2021.706229},
doi = {10.3389/fbioe.2021.706229},
ISSN = {2296-4185}
}
@article{Jia2020AttentionGCN,
title = {Attention-based Graph {ResNet} for Motor Intent Detection from Raw EEG signals},
author = {Jia, Shuyue and Hou, Yimin and Lun, Xiangmin and Lv, Jinglei},
journal = {arXiv preprint arXiv:2007.13484},
year = {2022}
}Our papers can be downloaded from:
- A Novel Approach of Decoding EEG Four-class Motor Imagery Tasks via Scout ESI and CNN
Codes and Tutorials for this work can be found here.
Overall Framework:
Proposed CNNs Architecture:
- GCNs-Net: A Graph Convolutional Neural Network Approach for Decoding Time-resolved EEG Motor Imagery Signals
Slides Presentation for this work can be found here.
- Deep Feature Mining via Attention-based BiLSTM-GCN for Human Motor Imagery Recognition
Slides Presentation for this work can be found here.
I think the following presentations would be helpful when you guys get engaged with Python and TensorFlow.
-
Python Environment Setting-up Tutorial download
-
Usage of Cloud Server and Setting-up Tutorial download
-
TensorFlow for Deep Learning Tutorial download
We always welcome contributions to help make EEG-DL Library better. If you would like to contribute or have any questions, please don't hesitate to email me at [email protected].
The library was created and open-sourced by Shuyue Jia, supervised by Prof. Yimin Hou, at the School of Automation Engineering, Northeast Electric Power University, Jilin, Jilin, China.