ELEKTRONN is a highly configurable toolkit for training 3D/2D CNNs and general Neural Networks.
It is written in Python 2 and based on Theano, which allows CUDA-enabled GPUs to significantly accelerate the pipeline.
The package includes a sophisticated training pipeline designed for classification/localisation tasks on 3D/2D images. Additionally, the toolkit offers training routines for tasks on non-image data.
ELEKTRONN 1.0 and 2.0 was originally created by Marius Killinger and Gregor Urban, with contributions by Sven Dorkenwald, under the supervision of Joergen Kornfeld at the Max Planck Institute For Medical Research in Heidelberg to solve connectomics tasks.
ELEKTRONN 1.0 and 2.0 has been superceded by the more flexible, PyTorch-based elektronn3 library. elektronn3 is actively developed and supported, so we encourage you to use it instead of ELEKTRONN 1.0/2.0.
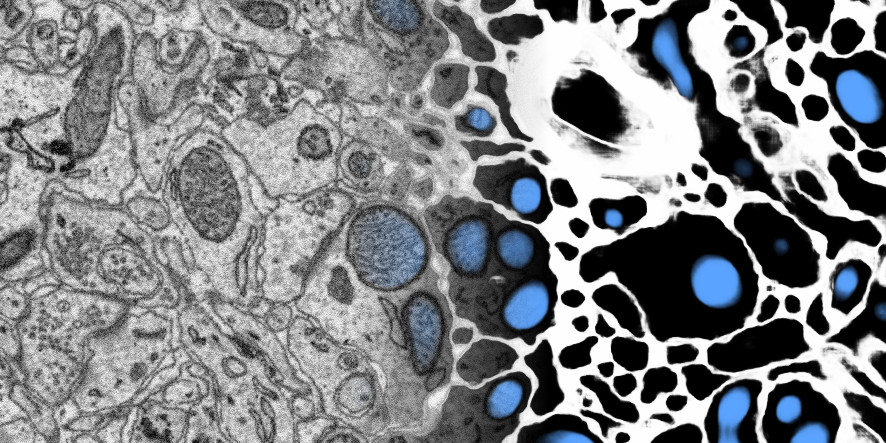
Membrane and mitochondria probability maps. Predicted with a CNN with recursive training. Data: zebra finch area X dataset j0126 by Jörgen Kornfeld.
$ elektronn-train MNIST_CNN_warp_config.py
This will download the MNIST data set and run a training defined in an example config file. The plots are saved to ~/CNN_Training/2D/MNIST_example_warp.
ELEKTRONN ├── doc # Documentation source files ├── elektronn │ ├── examples # Example scripts and config files │ ├── net # Neural network library code │ ├── scripts # Training script and profiling script │ ├── training # Training library code │ └── ... ├── LICENSE.rst ├── README.rst └── ...

