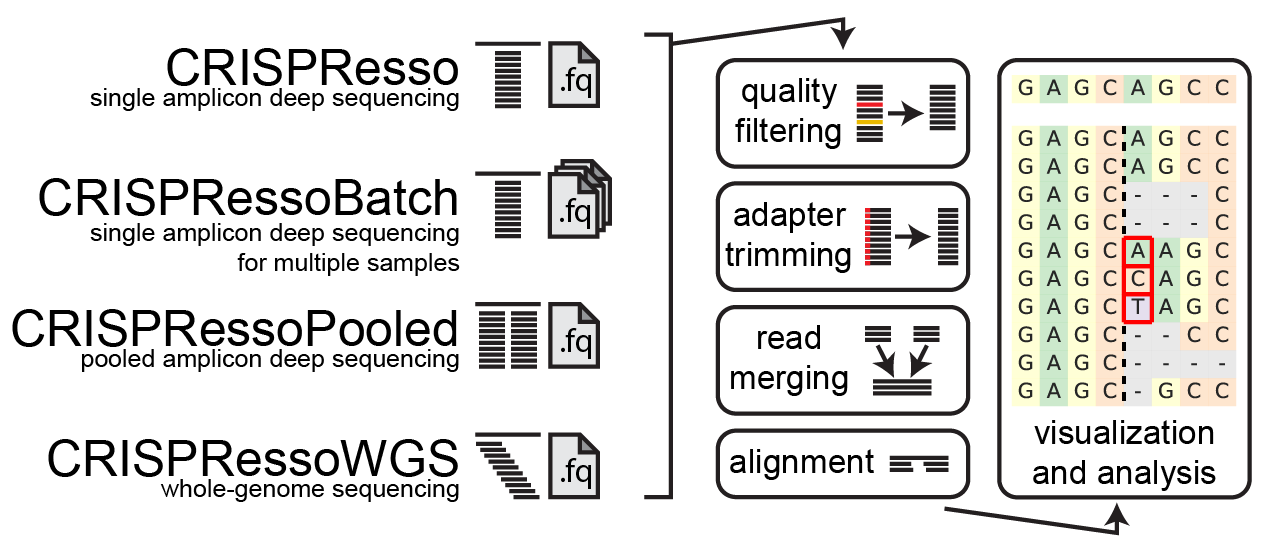
CRISPResso2 is a software pipeline designed to enable rapid and intuitive interpretation of genome editing experiments. A limited web implementation is available at: https://crispresso2.pinellolab.org/.
Briefly, CRISPResso2:
- aligns sequencing reads to a reference sequence
- quantifies insertions, mutations and deletions to determine whether a read is modified or unmodified by genome editing
- summarizes editing results in intuitive plots and datasets
CRISPResso2 can be used to analyze genome editing outcomes using cleaving nucleases (e.g. Cas9 or Cpf1) or noncleaving nucleases (e.g. base editors). The following operations can be automatically performed:
- filtering of low-quality reads
- adapter trimming
- alignment of reads to one or multiple reference sequences (in the case of multiple alleles)
- quantification of HDR and NHEJ outcomes (if the HDR sequence is provided)
- quantification frameshift/inframe mutations and identification affected splice sites (if an exon sequence is provided)
- visualization of the indel distribution and position (for cleaving nucleases)
- visualization of distribution and position of substitutions (for base editors)
- visualization of alleles and their frequencies
In addition, CRISPResso can be run as part of a larger tool suite:
- CRISPRessoBatch - for analyzing and comparing multiple experimental conditions at the same site
- CRISPRessoPooled - for analyzing multiple amplicons from a pooled amplicon sequencing experiment
- CRISPRessoWGS - for analyzing specific sites in whole-genome sequencing samples
- CRISPRessoCompare - for comparing editing between two samples (e.g., treated vs control)
- CRISPRessoAggregate - for aggregating results from previously-run CRISPResso analyses
Input reads are first filtered based on the quality score (phred33) in order to remove potentially false positive indels. The filtering based on the phred33 quality score can be modulated by adjusting the optimal parameters (see additional notes below).
Next, adapters are trimmed from the reads. If no adapter are present, select 'No Trimming' under the 'Trimming adapter' heading in the optional parameters. If reads contain adapter sequences that need to be trimmed, select the adapters used for trimming under the ‘Trimming adapter’ heading in the optional parameters. Possible adapters include Nextera PE, TruSeq3 PE, TruSeq3 SE, TruSeq2 PE, and TruSeq2 SE. The adapters are trimmed from the reads using fastp.
If paired-end reads are provided, reads are merged using fastp. This produces a single read for alignment to the amplicon sequence, and reduces sequencing errors that may be present at the end of sequencing reads.
The preprocessed reads are then aligned to the reference sequence with a global sequence alignment algorithm that takes into account our biological knowledge of nuclease function. If multiple alleles are present at the editing site, each allele can be passed to CRISPResso2 and sequenced reads will be assigned to the reference sequence or origin.
Finally, after analyzing the aligned reads, a set of informative graphs are generated, allowing for the quantification and visualization of the position and type of outcomes within the amplicon sequence.
CRISPResso2 introduces four key innovations for the analysis of genome editing data:
- Comprehensive analysis of sequencing data from base editors. We have added additional analysis and visualization capabilities especially for experiments using base editors.
- Allele specific quantification of heterozygous references. If the targeted editing region has more than one allele, reads arising from each allele can be deconvoluted.
- A novel biologically-informed alignment algorithm. This algorithm incorporates knowledge about the mutations produced by gene editing tools to create more biologically-likely alignments.
- Ultra-fast processing time.
CRISPResso2 can be installed using the conda package manager Bioconda, or it can be run using the Docker containerization system.
To install CRISPResso2 using Bioconda, download and install Anaconda Python, following the instructions at: https://www.anaconda.com/distribution/.
Open a terminal and type:
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
To install CRISPResso2 into the current conda environment, type:
conda install CRISPResso2
Alternatively, to create a new environment named crispresso2_env with CRISPResso2, type:
conda create -n crispresso2_env -c bioconda crispresso2
Activate your conda environment:
conda activate crispresso2_env
Verify that CRISPResso is installed using the command:
CRISPResso -h
If you would like to install CRISPResso using bioconda on a Mac with Apple silicon (aren't sure?), then there is a slight change you need to make. First, ensure that you have Rosetta installed. Next, you must tell bioconda to install the Intel versions of the packages. If you would like to do this system wide, which we recommend, run the command:
conda config --add subdirs osx-64Then you can proceed with the installation instructions above.
If you would like to use the Intel versions in a single environment, then run:
CONDA_SUBDIR=osx-64 conda create -n crispresso2_env -c bioconda crispresso2If you choose to use the CONDA_SUBDIR=osx-64 method, note that if you install additional packages into the environment you will need to add the CONDA_SUBDIR=osx-64 to the beginning of each command. Alternatively, you could set this environment variable in your shell, but we recommend to use the conda config --add subdirs osx-64 method because it is less error prone.
CRISPResso2 can be used via the Docker containerization system. This system allows CRISPResso2 to run on your system without configuring and installing additional packages. To run CRISPResso2, first download and install docker: https://docs.docker.com/engine/installation/
Next, Docker must be configured to access your hard drive and to run with sufficient memory. These parameters can be found in the Docker settings menu. To allow Docker to access your hard drive, select 'Shared Drives' and make sure your drive name is selected. To adjust the memory allocation, select the 'Advanced' tab and allocate at least 4G of memory.
To run CRISPResso2, make sure Docker is running, then open a command prompt (Mac) or Powershell (Windows). Change directories to the location where your data is, and run the following command:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso -h
The first time you run this command, it will download the Docker image. The -v parameter mounts the current directory to be accessible by CRISPResso2, and the -w parameter sets the CRISPResso2 working directory. As long as you are running the command from the directory containing your data, you should not change the Docker -v or -w parameters.
Additional parameters for CRISPResso2 as described below can be added to this command. For example,
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso -r1 sample.fastq.gz -a ATTAACCAAG
CRISPResso2 is designed be run on a single amplicon. For experiments involving multiple amplicons in the same fastq, see the instructions for CRISPRessoPooled or CRISPRessoWGS below.
CRISPResso2 requires only two parameters: input sequences in the form of fastq files (given by the --fastq_r1 and --fastq_r2) parameters, and the amplicon sequence to align to (given by the --amplicon_seq parameter). For example:
Using Bioconda:
CRISPResso --fastq_r1 reads.fastq.gz --amplicon_seq AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGGAGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCATCATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso --fastq_r1 reads.fastq.gz --amplicon_seq AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGGAGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCATCATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT
Guardrails automatically check the inputs and results of experiments against standardized values. The guardrail warnings that are triggered are printed in the commandline and at the top of generated reports. In order to turn off the guardrails, add the --disable_guardrails argument.
TotalReadsGuardrailChecks if the number of reads is lower than expected. (Default: 10000)OverallReadsAlignedGuardrailChecks if the number of aligned reads is lower than expected. (Default: 90% of the total reads)DisproportionateReadsAlignedGuardrailChecks if the number of reads aligned to an amplicon is higher or lower than expected proportionally. (Default: 30% more or less than expected)LowRatioOfModsInWindowToOutGuardrailChecks if the ratio of modifications inside to outside the quantification window is lower than expected. (Default: 0.01)HighRateOfModificationAtEndsGuardrailChecks if there is a high rate of modifications at the ends of the read. (Default: 0.01)HighRateOfSubstitutionsOutsideWindowGuardrailChecks if there is a high rate of substitutions outside of the quantification windows. (Default: 0.002)HighRateOfSubstitutionsGuardrailChecks if the proportion of substitutions to other modifications is higher than expected. (Default: 0.3)ShortSequenceGuardrailChecks if the provided sequences (both Amplicons and Guides) are shorter than expected. (Amplicon Default: 50, Guide Default: 19)LongAmpliconShortReadsGuardrailChecks if the provided amplicon is more than<value>times the average length of read. (Default: 1.5)
Download the test datasets nhej.r1.fastq.gz and nhej.r2.fastq.gz to your current directory. This is the first 25,000 sequences from a paired-end sequencing experiment. To analyze this experiment, run the command:
Using Bioconda:
CRISPResso --fastq_r1 nhej.r1.fastq.gz --fastq_r2 nhej.r2.fastq.gz --amplicon_seq AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGGAGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCATCATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT -n nhej
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso --fastq_r1 nhej.r1.fastq.gz --fastq_r2 nhej.r2.fastq.gz --amplicon_seq AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGGAGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCATCATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT -n nhej
This should produce a folder called 'CRISPResso_on_nhej'. Open the file called CRISPResso_on_nhej/CRISPResso2_report.html in a web browser, and you should see an output like this: CRISPResso2_report.html.
Download the test dataset allele_specific.fastq.gz to your current directory. This is the first 25,000 sequences from a editing experiment targeting one allele. To analyze this experiment, run the following command:
Using Bioconda:
CRISPResso --fastq_r1 allele_specific.fastq.gz --amplicon_seq CGAGAGCCGCAGCCATGAACGGCACAGAGGGCCCCAATTTTTATGTGCCCTTCTCCAACGTCACAGGCGTGGTGCGGAGCCACTTCGAGCAGCCGCAGTACTACCTGGCGGAACCATGGCAGTTCTCCATGCTGGCAGCGTACATGTTCCTGCTCATCGTGCTGGG,CGAGAGCCGCAGCCATGAACGGCACAGAGGGCCCCAATTTTTATGTGCCCTTCTCCAACGTCACAGGCGTGGTGCGGAGCCCCTTCGAGCAGCCGCAGTACTACCTGGCGGAACCATGGCAGTTCTCCATGCTGGCAGCGTACATGTTCCTGCTCATCGTGCTGGG --amplicon_name P23H,WT --guide_seq GTGCGGAGCCACTTCGAGCAGC
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso --fastq_r1 allele_specific.fastq.gz --amplicon_seq CGAGAGCCGCAGCCATGAACGGCACAGAGGGCCCCAATTTTTATGTGCCCTTCTCCAACGTCACAGGCGTGGTGCGGAGCCACTTCGAGCAGCCGCAGTACTACCTGGCGGAACCATGGCAGTTCTCCATGCTGGCAGCGTACATGTTCCTGCTCATCGTGCTGGG,CGAGAGCCGCAGCCATGAACGGCACAGAGGGCCCCAATTTTTATGTGCCCTTCTCCAACGTCACAGGCGTGGTGCGGAGCCCCTTCGAGCAGCCGCAGTACTACCTGGCGGAACCATGGCAGTTCTCCATGCTGGCAGCGTACATGTTCCTGCTCATCGTGCTGGG --amplicon_name P23H,WT --guide_seq GTGCGGAGCCACTTCGAGCAGC
This should produce a folder called 'CRISPResso_on_allele_specific'. Open the file called CRISPResso_on_allele_specific/CRISPResso2_report.html in a web browser, and you should see an output like this: CRISPResso2_report.html.
Download the test dataset base_editor.fastq.gz to your current directory. This is the first 25,000 sequences from an editing experiment performed at the EMX1 locus. To analyze this experiment, run the following command:
Using Bioconda:
CRISPResso --fastq_r1 base_editor.fastq.gz --amplicon_seq GGCCCCAGTGGCTGCTCTGGGGGCCTCCTGAGTTTCTCATCTGTGCCCCTCCCTCCCTGGCCCAGGTGAAGGTGTGGTTCCAGAACCGGAGGACAAAGTACAAACGGCAGAAGCTGGAGGAGGAAGGGCCTGAGTCCGAGCAGAAGAAGAAGGGCTCCCATCACATCAACCGGTGGCGCATTGCCACGAAGCAGGCCAATGGGGAGGACATCGATGTCACCTCCAATGACTAGGGTGG --guide_seq GAGTCCGAGCAGAAGAAGAA --quantification_window_size 10 --quantification_window_center -10 --base_editor_output
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso --fastq_r1 base_editor.fastq.gz --amplicon_seq GGCCCCAGTGGCTGCTCTGGGGGCCTCCTGAGTTTCTCATCTGTGCCCCTCCCTCCCTGGCCCAGGTGAAGGTGTGGTTCCAGAACCGGAGGACAAAGTACAAACGGCAGAAGCTGGAGGAGGAAGGGCCTGAGTCCGAGCAGAAGAAGAAGGGCTCCCATCACATCAACCGGTGGCGCATTGCCACGAAGCAGGCCAATGGGGAGGACATCGATGTCACCTCCAATGACTAGGGTGG --guide_seq GAGTCCGAGCAGAAGAAGAA --quantification_window_size 10 --quantification_window_center -10 --base_editor_output
This should produce a folder called 'CRISPResso_on_base_editor'. Open the file called CRISPResso_on_base_editor/CRISPResso2_report.html in a web browser, and you should see an output like this: CRISPResso2_report.html.
-h or --help: show a help message and exit.
-r1 or --fastq_r1: The first fastq file.
-r2 or --fastq_r2 FASTQ_R2: The second fastq file for paired end reads.
-a or --amplicon_seq: The amplicon sequence used for the experiment.
-an or --amplicon_name: A name for the reference amplicon can be given. If multiple amplicons are given, multiple names can be specified here. Because amplicon names are used as output filename prefixes, amplicon names are truncated to 21bp unless the parameter --suppress_amplicon_name_truncation is set. (default: Reference)
-g or --guide_seq: sgRNA sequence, if more than one, please separate by commas. Note that the sgRNA needs to be input as the guide RNA sequence (usually 20 nt) immediately adjacent to but not including the PAM sequence (5' of NGG for SpCas9). If the PAM is found on the opposite strand with respect to the Amplicon Sequence, ensure the sgRNA sequence is also found on the opposite strand. The CRISPResso convention is to depict the expected cleavage position using the value of the parameter '--quantification_window_center' nucleotides from the 3' end of the guide. In addition, the use of alternate nucleases besides SpCas9 is supported. For example, if using the Cpf1 system, enter the sequence (usually 20 nt) immediately 3' of the PAM sequence and explicitly set the '--cleavage_offset' parameter to 1, since the default setting of -3 is suitable only for SpCas9. (default: )
-e or --expected_hdr_amplicon_seq: Amplicon sequence expected after HDR. The expected HDR amplicon sequence can be provided to quantify the number of reads showing a successful HDR repair. Note that the entire amplicon sequence must be provided, not just the donor template. CRISPResso2 will quantify identified instances of NHEJ, HDR, or mixed editing events. (default: )
-c or --coding_seq: Subsequence/s of the amplicon sequence covering one or more coding sequences for frameshift analysis. Sequences of exons within the amplicon sequence can be provided to enable frameshift analysis and splice site analysis by CRISPResso2. If more than one (for example, split by intron/s), please separate by commas. Users should provide the subsequences of the reference amplicon sequence that correspond to coding sequences (not the whole exon sequence(s)!). (default: )
-gn or --guide_name: sgRNA names, if more than one, please separate by commas. (default: sgRNA)
-fg or --flexiguide: sgRNA sequence (flexible). The flexiguide sequence will be aligned to the amplicon sequence(s), as long as the guide sequence has homology as set by --flexiguide_homology. (default: '')
-fh or --flexiguide_homology: flexiguides will yield guides in amplicons with at least this homology to the flexiguide sequence (default:80 meaning 80% homology is required)
-fgn or --flexiguide_name: Names for the flexiguides, similar to --guide_name. (default: '')
--discard_guide_positions_overhanging_amplicon_edge: If set, for guides that align to multiple positions, guide positions will be discarded if plotting around those regions would included bp that extend beyond the end of the amplicon. (default: False)
--split_interleaved_input: Splits a single fastq file containing paired end reads in two files before running CRISPResso (default: False)
-q or --min_average_read_quality: Minimum average quality score (phred33) to keep a read (default: 0)
-s or --min_single_bp_quality: Minimum single bp score (phred33) to keep a read (default: 0)
--min_bp_quality_or_N: Bases with a quality score (phred33) less than this value will be set to "N" (default: 0)
--trim_sequences: Enable the trimming of Illumina adapters with fastp (default: False)
--fastp_options_string: Override options for fastp, e.g. --length_required 70 --umi
--min_paired_end_reads_overlap: Parameter for the fastp read merging step. Minimum required overlap length between two reads to provide a confident overlap. (default: 10)
-w or --quantification_window_size or --window_around_sgrna: Defines the size (in bp) of the quantification window extending from the position specified by the "--cleavage_offset" or "--quantification_window_center" parameter in relation to the provided guide RNA sequence(s) (--sgRNA). Mutations within this number of bp from the quantification window center are used in classifying reads as modified or unmodified. A value of 0 disables this window and indels in the entire amplicon are considered. Default is 1, 1bp on each side of the cleavage position for a total length of 2bp. (default: 1)
-wc or --quantification_window_center or --cleavage_offset: Center of quantification window to use within respect to the 3' end of the provided sgRNA sequence. Remember that the sgRNA sequence must be entered without the PAM. For cleaving nucleases, this is the predicted cleavage position. The default is -3 and is suitable for the Cas9 system. For alternate nucleases, other cleavage offsets may be appropriate, for example, if using Cpf1 this parameter would be set to 1. For base editors, this could be set to -17. (default: -3)
-qwc or --quantification_window_coordinates: Bp positions in the amplicon sequence specifying the quantification window. This parameter overrides values of the "--quantification_window_center", "-- cleavage_offset", "--window_around_sgrna" or "-- window_around_sgrna" values. Any indels/substitutions outside this window are excluded. Indexes are 0-based, meaning that the first nucleotide is position 0. Ranges are separated by the dash sign like "start-stop", and multiple ranges can be separated by the underscore (_). A value of 0 disables this filter. (can be comma-separated list of values, corresponding to amplicon sequences given in --amplicon_seq e.g. 5-10,5-10_20-30 would specify the 5th-10th bp in the first reference and the 5th-10th and 20th-30th bp in the second reference). Note that if there are multiple amplicons provided, and only one quantification window coordinate is provided, the same quantification window will be used for all amplicons and be adjusted to account for insertions/deletions. (default: None)
--exclude_bp_from_left: Exclude bp from the left side of the amplicon sequence for the quantification of the indels (default: 15)
--exclude_bp_from_right: Exclude bp from the right side of the amplicon sequence for the quantification of the indels (default: 15)
--ignore_substitutions: Ignore substitutions events for the quantification and visualization (default: False)
--ignore_insertions: Ignore insertions events for the quantification and visualization (default: False)
--ignore_deletions: Ignore deletions events for the quantification and visualization (default: False)
--discard_indel_reads: Discard reads with indels in the quantification window from analysis (default: False)
-amas or --amplicon_min_alignment_score: Amplicon Minimum Alignment Score; score between 0 and 100; sequences must have at least this homology score with the amplicon to be aligned (can be comma-separated list of multiple scores, corresponding to amplicon sequences given in --amplicon_seq) After reads are aligned to a reference sequence, the homology is calculated as the number of bp they have in common. If the aligned read has a homology less than this parameter, it is discarded. This is useful for filtering erroneous reads that do not align to the target amplicon, for example arising from alternate primer locations. (default: 60)
--default_min_aln_score or --min_identity_score: Default minimum homology score for a read to align to a reference amplicon (default: 60)
--expand_ambiguous_alignments: If more than one reference amplicon is given, reads that align to multiple reference amplicons will count equally toward each amplicon. Default behavior is to exclude ambiguous alignments. (default: False)
--assign_ambiguous_alignments_to_first_reference: If more than one reference amplicon is given, ambiguous reads that align with the same score to multiple amplicons will be assigned to the first amplicon. Default behavior is to exclude ambiguous alignments. (default: False)
--needleman_wunsch_gap_open: Gap open option for Needleman-Wunsch alignment (default: -20)
--needleman_wunsch_gap_extend: Gap extend option for Needleman-Wunsch alignment (default: -2)
--needleman_wunsch_gap_incentive: Gap incentive value for inserting indels at cut sites (default: 1)
--needleman_wunsch_aln_matrix_loc: Location of the matrix specifying substitution scores in the NCBI format (see ftp://ftp.ncbi.nih.gov/blast/matrices/) (default: EDNAFULL)
--base_editor_output: Outputs plots and tables to aid in analysis of base editor studies. If base editor output is selected, plots showing the frequency of substitutions in the quantification window are generated. The target and result bases can also be set to measure the rate of on-target conversion at bases in the quantification window. (default: False)
--conversion_nuc_from: For base editor plots, this is the nucleotide targeted by the base editor (default: C)
--conversion_nuc_to: For base editor plots, this is the nucleotide produced by the base editor (default: T)
--prime_editing_pegRNA_spacer_seq: pegRNA spacer sgRNA sequence used in prime editing. The spacer should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the given sequence. (default: )
--prime_editing_pegRNA_extension_seq: Extension sequence used in prime editing. The sequence should be given in the RNA 5'->3' order, such that the sequence starts with the RT template including the edit, followed by the Primer-binding site (PBS). (default: )
--prime_editing_pegRNA_extension_quantification_window_size: Quantification window size (in bp) at flap site for measuring modifications anchored at the right side of the extension sequence. Similar to the --quantification_window parameter, the total length of the quantification window will be 2x this parameter. Default: 5bp (10bp total window size) (default: 5)
--prime_editing_pegRNA_scaffold_seq: If given, reads containing any of this scaffold sequence before extension sequence (provided by --prime_editing_extension_seq) will be classified as 'Scaffold-incorporated'. The sequence should be given in the 5'->3' order such that the RT template directly follows this sequence. A common value ends with 'GGCACCGAGUCGGUGC'. (default: )
--prime_editing_pegRNA_scaffold_min_match_length: Minimum number of bases matching scaffold sequence for the read to be counted as 'Scaffold-incorporated'. If the scaffold sequence matches the reference sequence at the incorporation site, the minimum number of bases to match will be minimally increased (beyond this parameter) to disambiguate between prime-edited and scaffold-incorporated sequences. (default: 1)
--prime_editing_nicking_guide_seq: Nicking sgRNA sequence used in prime editing. The sgRNA should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the sequence (default: )
--prime_editing_override_prime_edited_ref_seq: If given, this sequence will be used as the prime-edited reference sequence. This may be useful if the prime-edited reference sequence has large indels or the algorithm cannot otherwise infer the correct reference sequence. (default='')
--plot_histogram_outliers: If set, all values will be shown on histograms. By default (if unset), histogram ranges are limited to plotting data within the 99 percentile. (default: False)
--plot_window_size or --offset_around_cut_to_plot: Defines the size of the window extending from the quantification window center to plot. Nucleotides within plot_window_size of the quantification_window_center for each guide are plotted. (default: 20)
--min_frequency_alleles_around_cut_to_plot: Minimum % reads required to report an allele in the alleles table plot. This parameter only affects plotting. All alleles will be reported in data files. (default: 0.2 (i.e. 0.2%))
--max_rows_alleles_around_cut_to_plot: Maximum number of rows to report in the alleles table plot. (default: 50)
--expand_allele_plots_by_quantification: If set, alleles with different modifications in the quantification window (but not necessarily in the plotting window (e.g. for another sgRNA)) are plotted on separate lines, even though they may have the same apparent sequence. To force the allele plot and the allele table to be the same, set this parameter. If unset, all alleles with the same sequence will be collapsed into one row. (default: False)
--allele_plot_pcts_only_for_assigned_reference: If set, in the allele plots, the percentages will show the percentage as a percent of reads aligned to the assigned reference. Default behavior is to show percentage as a percent of all reads. (default: False)
--annotate_wildtype_allele: Wildtype alleles in the allele table plots will be marked with this string (e.g. **). (default: )
--file_prefix: File prefix for output plots and tables (default: )
-n or --name: Output name of the report (default: the names is obtained from the filename of the fastq file/s used in input) (default: )
-o or --output_folder: Output folder to use for the analysis (default: current folder)
--write_detailed_allele_table: If set, a detailed allele table will be written including alignment scores for each read sequence. (default: False)
--suppress_amplicon_name_truncation: If set, amplicon names will not be truncated when creating output filename prefixes. If not set, amplicon names longer than 21 characters will be truncated when creating filename prefixes. (default: False)
--fastq_output: If set, a fastq file with annotations for each read will be produced. (default: False)
--bam_output': If set, a bam file with alignments for each read will be produced. Setting this parameter will produce a file called 'CRISPResso_output.bam' with the alignments in bam format. If the bowtie2_index is provided, alignments will be reported in reference to that genome. If the bowtie2_index is not provided, alignments will be reported in reference to a custom reference created by the amplicon sequence(s) and written to the file 'CRISPResso_output.fa'. (default: False)
-x or --bowtie2_index: Basename of Bowtie2 index for the reference genome. Optionally used in the creation of a bam file. See bam_output. (default: '')
--keep_intermediate: Keep all the intermediate files (default: False)
--dump: Dump numpy arrays and pandas dataframes to file for debugging purposes (default: False)
--crispresso1_mode: Output as in CRISPResso1. In particular, if this flag is set, the old output files 'Mapping_statistics.txt', and 'Quantification_of_editing_frequency.txt' are created, and the new files 'nucleotide_frequency_table.txt' and 'substitution_frequency_table.txt' and figure 2a and 2b are suppressed, and the files 'selected_nucleotide_percentage_table.txt' are not produced when the flag --base_editor_output is set (default: False)
--suppress_report: Suppress output report, plots output as .pdf only (not .png) (default: False)
--suppress_plots: Suppress output plots (default: False)
--place_report_in_output_folder: If true, report will be written inside the CRISPResso output folder. By default, the report will be written one directory up from the report output. (default: False)
--zip_output: If true, the output folder will be zipped upon completion. If --zip_output is true --place_report_in_output_folder should be true otherwise --place_report_in_output_folder is automatically set to true as well. (default: False)
--auto: Infer amplicon sequence from most common reads (default: False)
--dsODN: dsODN sequence -- Reads containing the dsODN are labeled and quantified. (default: '')
--debug: Show debug messages (default: False)
-v or --verbosity: Verbosity level of output to the console (1-4), 4 is the most verbose. If parameter --debug is set --verbosity is overridden and set to 4. (default=3)
--no_rerun: Don't rerun CRISPResso2 if a run using the same parameters has already been finished. (default: False)
--bam_input BAM_INPUT: Aligned reads for processing in bam format. This parameter can be given instead of fastq_r1 to specify that reads are to be taken from this bam file. An output bam is produced that contains an additional field with CRISPResso2 information. (default: )
--bam_chr_loc BAM_CHR_LOC: Chromosome location in bam for reads to process. For example: "chr1:50-100" or "chrX". (default: )
--disable_guardrails: Don't show the guardrail warnings.
The output of CRISPResso2 consists of a set of informative graphs that allow for the quantification and visualization of the position and type of outcomes within an amplicon sequence.
CRISPResso2_report.html is a summary report that can be viewed in a web browser containing all of the output plots and summary statistics.
Alleles_frequency_table.zip can be unzipped to a tab-separated text file that shows all reads and alignments to references. The first column shows the aligned sequence of the sequenced read. The second column shows the aligned sequence of the reference sequence. Gaps in each of these columns represent insertions and deletions. The next column 'Reference_Name' shows the name of the reference that the read aligned to. The fourth column, 'Read_Status' shows whether the read was modified or unmodified. The fifth through seventh columns ('n_deleted', 'n_inserted', 'n_substituted') show the number of bases deleted, inserted, and substituted as compared to the reference sequence. The eighth column shows the number of reads having that sequence, and the ninth column shows the percentage of all reads having that sequence.
CRISPResso_mapping_statistics.txt is a tab-delimited text file showing the number of reads in the input ('READS IN INPUTS') the number of reads after filtering, trimming and merging (READS AFTER PREPROCESSING), the number of reads aligned (READS ALIGNED) and the number of reads for which the alignment had to be computed vs read from cache.
CRISPResso_quantification_of_editing_frequency.txt is a tab-delimited text file showing the number of reads aligning to each reference amplicon, as well as the status (modified/unmodified, number of insertions, deletions, and/or substitutions) of those reads.
CRISPResso_RUNNING_LOG.txt is a text file and shows a log of the CRISPResso run.
CRISPResso2_info.json can be read by other CRISPResso tools and contains information about the run and results.
The remainder of the files are produced for each amplicon, and each file is prefixed by the name of the amplicon if more than one amplicon is given.
Alleles_frequency_table_around_sgRNA_NNNNN.txt is a tab-separated text file that shows alleles and alignments to the specified reference for a subsequence around the sgRNA (here, shown by 'NNNNN'). This data report is produced for each amplicon when a guide is found in the amplicon sequence. A report is generated for each guide. The number of nucleotides shown in this report can be modified by changing the --plot_window_size parameter.
Substitution_frequency_table_around_sgRNA_NNNNN.txt is a tab-separated text file that shows the frequency of substitutions in the amplicon sequence around the sgRNA (here, shown by 'NNNNN'). The first row shows the reference sequence. The following rows show the number of substitutions to each base. For example, the first numeric value in the second row (marked ‘A’) shows the number of bases that have a substitution resulting in an A at the first basepair of the amplicon sequence. The number of unmodified bases at each position is now shown in this table (because they aren’t substitutions). Thus, if the first basepair of the amplicon sequence is an A, the first value in the first row will show 0. A report is generated for each guide. The number of nucleotides shown in this report can be modified by changing the --plot_window_size parameter.
Substitution_frequency_table.txt is a tab-separated text file that shows the frequency of substitutions in the amplicon sequence across the entire amplicon. The first row shows the reference sequence. The following rows show the number of substitutions to each base. For example, the first numeric value in the second row (marked ‘A’) shows the number of bases that have a substitution resulting in an A at the first basepair of the amplicon sequence. The number of unmodified bases at each position is now shown in this table (because they aren’t substitutions). Thus, if the first basepair of the AMPLICON sequence is an A, the first value in the first row will show 0.
Insertion_histogram.txt is a tab-separated text file that shows a histogram of the insertion sizes in the amplicon sequence in the quantification window. Insertions outside of the quantification window are not included. The ins_size column shows the insertion length, and the fq column shows the number of reads having that insertion size.
Deletion_histogram.txt is a tab-separated text file that shows a histogram of the deletion sizes in the amplicon sequence in the quantification window. Deletions outside of the quantification window are not included. The del_size column shows length of the deletion, and the fq column shows the number of reads having that number of substitutions.
Substitution_histogram.txt is a tab-separated text file that shows a histogram of the number of substitutions in the amplicon sequence in the quantification window. Substitutions outside of the quantification window are not included. The sub_count column shows the number of substitutions, and the fq column shows the number of reads having that number of substitutions.
Effect_vector_insertion.txt is a tab-separated text file with a one-row header that shows the percentage of reads with an insertion at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a insertion at that location.
Effect_vector_deletion.txt is a tab-separated text file with a one-row header that shows the percentage of reads with a deletion at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a deletion at that location.
Effect_vector_substitution.txt is a tab-separated text file with a one-row header that shows the percentage of reads with a substitution at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a substitution at that location.
Effect_vector_combined.txt is a tab-separated text file with a one-row header that shows the percentage of reads with any modification (insertion, deletion, or substitution) at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a modification at that location.
Modification_count_vectors.txt is a tab-separated file showing the number of modifications for each position in the amplicon. The first row shows the amplicon sequence, and successive rows show the number of reads with insertions (row 2), insertions_left (row 3), deletions (row 4), substitutions (row 5) and the sum of all modifications (row 6). Additionally, the last row shows the number of reads aligned.
If an insertion occurs between bases 5 and 6, the insertions vector will be incremented at bases 5 and 6. However, the insertions_left vector will only be incremented at base 5 so the sum of the insertions_left row represents an accurate count of the number of insertions, whereas the sum of the insertions row will yield twice the number of insertions.
Quantification_window_modification_count_vectors.txt is a tab-separated file showing the number of modifications for positions in the quantification window of the amplicon. The first row shows the amplicon sequence in the quantification window, and successive rows show the number of reads with insertions (row 2), insertions_left (row 3), deletions (row 4), substitutions (row 5) and the sum of all modifications (row 6). Additionally, the last row shows the number of reads aligned.
Nucleotide_frequency_table.txt is a tab-separated file showing the number of each residue at each position in the amplicon. The first row shows the amplicon sequence, and successive rows show the number of reads with an A (row 2), C (row 3), G (row 4), T (row 5), N (row 6), or a deletion (-) (row 7) at each position.
Quantification_window_nucleotide_frequency_table.txt is a tab-separated file showing the number of each residue at positions in the quantification window of the amplicon. The first row shows the amplicon sequence in the quantification window, and successive rows show the number of reads with an A (row 2), C (row 3), G (row 4), T (row 5), N (row 6), or a deletion (-) (row 7) at each position.
Nucleotide_percentage_table.txt is a tab-separated file showing the percentage of each residue at each position in the amplicon. The first row shows the amplicon sequence, and successive rows show the percentage of reads with an A (row 2), C (row 3), G (row 4), T (row 5), N (row 6), or a deletion (-) (row 7) at each position.
Quantification_window_nucleotide_percentage_table.txt is a tab-separated file showing the percentage of each residue at positions in the quantification window of the amplicon. The first row shows the amplicon sequence in the quantification window, and successive rows show the percentage of reads with an A (row 2), C (row 3), G (row 4), T (row 5), N (row 6), or a deletion (-) (row 7) at each position.
The following report files are produced when the base editor mode is enabled:
Selected_nucleotide_percentage_table_around_sgRNA_NNNNN.txt is a tab-separated text file that shows the percentage of each base at selected nucleotides in the amplicon sequence around the sgRNA (here, shown by 'NNNNN'). If the base editing experiment targets cytosines (as set by the --base_editor_from parameter), each C in the quantification window will be numbered (e.g. C5 represents the cytosine at the 5th position in the selected nucleotides). The percentage of each base at these selected target cytosines is reported, with the first row showing the numbered cytosines, and the remainder of the rows showing the percentage of each nucleotide present at these locations. This file shows nucleotides within '--plot_window_size' bp of the position specified by the parameter '--quantification_window_center' relative to the 3' end of each guide.
Selected_nucleotide_frequency_table_around_sgRNA_NNNNN.txt is a tab-separated text file that shows the frequency of each base at selected nucleotides in the amplicon sequence around the sgRNA (here, shown by 'NNNNN'). If the base editing experiment targets cytosines (as set by the --base_editor_from parameter), each C in the quantification window will be numbered (e.g. C5 represents the cytosine at the 5th position in the selected nucleotides). The frequency of each base at these selected target cytosines is reported, with the first row showing the numbered cytosines, and the remainder of the rows showing the frequency of each nucleotide present at these locations. This file shows nucleotides within '--plot_window_size' bp of the position specified by the parameter '--quantification_window_center' relative to the 3' end of each guide.
The following report files are produced when the amplicon contains a coding sequence:
Frameshift_analysis.txt is a text file describing the number of noncoding, in-frame, and frameshift mutations. This report file is produced when the amplicon contains a coding sequence.
Splice_sites_analysis.txt is a text file describing the number of splicing sites that are unmodified and modified. This file report is produced when the amplicon contains a coding sequence.
Effect_vector_insertion_noncoding.txt is a tab-separated text file with a one-row header that shows the percentage of reads with a noncoding insertion at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a noncoding insertion at that location. This report file is produced when amplicon contains a coding sequence.
Effect_vector_deletion_noncoding.txt is a tab-separated text file with a one-row header that shows the percentage of reads with a noncoding deletion at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a noncoding deletion at that location. This report file is produced when amplicon contains a coding sequence.
Effect_vector_substitution_noncoding.txt is a tab-separated text file with a one-row header that shows the percentage of reads with a noncoding substitution at each base in the reference sequence. The first column shows the 1-based position of the amplicon, and the second column shows the percentage of reads with a noncoding substitution at that location. This report file is produced when amplicon contains a coding sequence.
Please check that your input file(s) are in FASTQ format (compressed fastq.gz also accepted).
If you get an empty report, please double check that your amplicon sequence is correct and in the correct orientation. It can be helpful to inspect the first few lines of your FASTQ file - the start of the amplicon sequence should match the start of your sequences. If not, check to see if the files are trimmed (see point below).
It is important to determine whether your reads are trimmed or not. CRISPResso2 assumes that the reads ARE ALREADY TRIMMED! If reads are not already trimmed, select the adapters used for trimming under the ‘Trimming Adapter’ heading under the ‘Optional Parameters’. This is FUNDAMENTAL to CRISPResso analysis. Failure to trim adaptors may result in false positives. This will result in a report where you will observe an unrealistic 100% modified alleles and a sharp peak at the edges of the reference amplicon in figure 4.
The quality filter assumes that your reads uses the Phred33 scale, and it should be adjusted for each user’s specific application. A reasonable value for this parameter is 30.
If your amplicon sequence is longer than your sequenced read length, the R1 and R2 reads should overlap by at least 10bp. For example, if you sequence using 150bp reads, the maximum amplicon length should be 290 bp.
Especially in repetitive regions, multiple alignments may have the best score. If you want to investigate alternate best-scoring alignments, you can view all alignments using this tool: http://rna.informatik.uni-freiburg.de/Teaching/index.jsp?toolName=Gotoh. As input, sequences from the 'Alleles_frequency_table.txt' can be used. Specifically, for a given row, the value in the 'Aligned_Sequence' should be entered into the 'Sequence a' box after removing any dashes, and the value in the 'Reference_Sequence' should be entered into the 'Sequence b' box after removing any dashes. The alternate alignments can be selected in the 'Results' panel in the Output section.
CRISPResso2 can be run for many fastqs (CRISPRessoBatch), for many amplicons in the same fastq (CRISPRessoPooled), or for whole-genome sequencing (CRISPRessoWGS).
CRISPRessoBatch allows users to specify input files and other command line arguments in a single file, and then to run CRISPResso2 analysis on each file in parallel. Samples for which the amplicon and guide sequences are the same will be compared between batches, producing useful summary tables and coomparison plots.
This flexible utility adds four additional parameters:
--batch_settings: This parameter specifies the tab-separated batch file. The batch file consists of a header line listing the parameters specified, and then one line for each sample describing the parameters for that sample. Each of the parameters for CRISPResso2 given above can be specified for each sample. When CRISPRessoBatch is run, additional parameters can be specified that will be applied to all of the samples listed in the batch file. An example batch file looks like:
name fastq_r1
sample1 sample1.fq
sample2 sample2.fq
sample3 sample3.fq
--skip_failed: If any sample fails, CRISPRessoBatch will exit without completion. However, if this parameter is specified, CRISPressoBatch will continue and only summarize the statistics of the successfully-completed runs.
-p or --n_processes: This specifies the number of processes to use for quantification. (default: 1)
-bo or --batch_output_folder: Directory where batch analysis output will be stored.
CRISPRessoBatch outputs several summary files and plots:
CRISPRessoBatch_quantification_of_editing_frequency shows the number of reads that were modified for each amplicon in each sample.
CRISPRessoBatch_mapping_statistics.txt aggregates the read mapping data from each sample.
For each amplicon, the following files are produced with the name of the amplicon as the filename prefix:
NUCLEOTIDE_FREQUENCY_SUMMARY.txt and NUCLEOTIDE_PERCENTAGE_SUMMARY.txt aggregate the nucleotide counts and percentages at each position in the amplicon for each sample.
MODIFICATION_FREQUENCY_SUMMARY.txt and MODIFICATION_PERCENTAGE_SUMMARY.txt aggregate the modification frequency and percentage at each position in the amplicon for each sample.
Download the test dataset files SRR3305543.fastq.gz, SRR3305544.fastq.gz, SRR3305545.fastq.gz, and SRR3305546.fastq.gz to your current directory. These are files are the first 25,000 sequences from an editing experiment performed on several base editors. Also include a batch file that lists these files and the sample names: batch.batch To analyze this experiment, run the following command:
Using Bioconda:
CRISPRessoBatch --batch_settings batch.batch --amplicon_seq CATTGCAGAGAGGCGTATCATTTCGCGGATGTTCCAATCAGTACGCAGAGAGTCGCCGTCTCCAAGGTGAAAGCGGAAGTAGGGCCTTCGCGCACCTCATGGAATCCCTTCTGCAGCACCTGGATCGCTTTTCCGAGCTTCTGGCGGTCTCAAGCACTACCTACGTCAGCACCTGGGACCCC -p 4 --base_editor_output -g GGAATCCCTTCTGCAGCACC -wc -10 -w 20
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoBatch --batch_settings batch.batch --amplicon_seq CATTGCAGAGAGGCGTATCATTTCGCGGATGTTCCAATCAGTACGCAGAGAGTCGCCGTCTCCAAGGTGAAAGCGGAAGTAGGGCCTTCGCGCACCTCATGGAATCCCTTCTGCAGCACCTGGATCGCTTTTCCGAGCTTCTGGCGGTCTCAAGCACTACCTACGTCAGCACCTGGGACCCC -p 4 --base_editor_output -g GGAATCCCTTCTGCAGCACC -wc -10 -w 20
This should produce a folder called 'CRISPRessoBatch_on_batch'. Open the file called CRISPRessoBatch_on_batch/CRISPResso2Batch_report.html in a web browser, and you should see an output like this: CRISPResso2Batch_report.html.
CRISPRessoPooled is a utility to analyze and quantify targeted sequencing CRISPR/Cas9 experiments involving pooled amplicon sequencing libraries. One common experimental strategy is to pool multiple amplicons (e.g. a single on-target site plus a set of potential off-target sites) into a single deep sequencing reaction (briefly, genomic DNA samples for pooled applications can be prepared by first amplifying the target regions for each gene/target of interest with regions of 150-400bp depending on the desired coverage. In a second round of PCR, with minimized cycle numbers, barcode and adaptors are added. With optimization, these two rounds of PCR can be merged into a single reaction. These reactions are then quantified, normalized, pooled, and undergo quality control before being sequenced). CRISPRessoPooled demultiplexes reads from multiple amplicons and runs the CRISPResso utility with appropriate reads for each amplicon separately.
This tool can run in 3 different modes:
Amplicons mode: Given a set of amplicon sequences, in this mode the tool demultiplexes the reads, aligning each read to the amplicon with best alignment, and creates separate compressed FASTQ files, one for each amplicon. Reads that do not align to any amplicon are discarded. After this preprocessing, CRISPResso is run for each FASTQ file, and separated reports are generated, one for each amplicon.
To run the tool in this mode the user must provide:
-
Paired-end reads (two files) or single-end reads (single file) in FASTQ format (fastq.gz files are also accepted)
-
A description file containing the amplicon sequences used to enrich regions in the genome and some additional information. In particular, this file, is a tab delimited text file with up to 12 columns (first 2 columns required):
-
AMPLICON_NAME: an identifier for the amplicon (must be unique).
-
AMPLICON_SEQUENCE: amplicon sequence used in the design of the experiment.
-
sgRNA_SEQUENCE (OPTIONAL): sgRNA sequence used for this amplicon without the PAM sequence. If not available, enter NA.
-
EXPECTED_AMPLICON_AFTER_HDR (OPTIONAL): expected amplicon sequence in case of HDR. If more than one, separate by commas and not spaces. If not available, enter NA.
-
CODING_SEQUENCE (OPTIONAL): Subsequence(s) of the amplicon corresponding to coding sequences. If more than one, separate by commas and not spaces. If not available, enter NA.
-
PRIME_EDITING_PEGRNA_SPACER_SEQ (OPTIONAL): pegRNA spacer sgRNA sequence used in prime editing. The spacer should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the given sequence. If not available, enter NA.
-
PRIME_EDITING_NICKING_GUIDE_SEQ (OPTIONAL): Nicking sgRNA sequence used in prime editing. The sgRNA should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the sequence. If not available, enter NA.
-
PRIME_EDITING_PEGRNA_EXTENSION_SEQ (OPTIONAL): Extension sequence used in prime editing. The sequence should be given in the RNA 5'->3' order, such that the sequence starts with the RT template including the edit, followed by the Primer-binding site (PBS). If not available, enter NA.
-
PRIME_EDITING_PEGRNA_SCAFFOLD_SEQ (OPTIONAL): If given, reads containing any of this scaffold sequence before extension sequence (provided by --primeediting_extension_seq) will be classified as 'Scaffold-incorporated'. The sequence should be given in the 5'->3' order such that the RT template directly follows this sequence. A common value ends with 'GGCACCGAGUCGGUGC'. If not available, enter _NA.
-
PRIME_EDITING_PEGRNA_SCAFFOLD_MIN_MATCH_LENGTH (OPTIONAL): Minimum number of bases matching scaffold sequence for the read to be counted as 'Scaffold-incorporated'. If the scaffold sequence matches the reference sequence at the incorporation site, the minimum number of bases to match will be minimally increased (beyond this parameter) to disambiguate between prime-edited and scaffold-incorporated sequences. If not available, enter NA.
-
PRIME_EDITING_OVERRIDE_PRIME_EDITED_REF_SEQ (OPTIONAL):If given, this sequence will be used as the prime-edited reference sequence. This may be useful if the prime-edited reference sequence has large indels or the algorithm cannot otherwise infer the correct reference sequence. If not available, enter NA.
-
QWC or QUANTIFICATION_WINDOW_COORDINATES (OPTIONAL): Bp positions in the amplicon sequence specifying the quantification window. Any indels/substitutions outside this window are excluded. Indexes are 0-based, meaning that the first nucleotide is position 0. Ranges are separated by the dash sign like "start-stop", and multiple ranges can be separated by the underscore (_). A value of 0 disables this filter. If not available, enter NA.
-
W or QUANTIFICATION_WINDOW_SIZE (OPTIONAL): Defines the size (in bp) of the quantification window extending from the position specified by the "--cleavage_offset" or "--quantification_window_center" parameter in relation to the provided guide RNA sequence(s) (--sgRNA). Mutations within this number of bp from the quantification window center are used in classifying reads as modified or unmodified. A value of 0 disables this window and indels in the entire amplicon are considered. Default is 1, 1bp on each side of the cleavage position for a total length of 2bp. (default: 1) If not available, enter _NA.*
-
WC or QUANTIFICATION_WINDOW_CENTER (OPTIONAL): Center of quantification window to use within respect to the 3' end of the provided sgRNA sequence. Remember that the sgRNA sequence must be entered without the PAM. For cleaving nucleases, this is the predicted cleavage position. The default is -3 and is suitable for the Cas9 system. For alternate nucleases, other cleavage offsets may be appropriate, for example, if using Cpf1 this parameter would be set to 1. For base editors, this could be set to -17. (default: -3) If not available, enter NA.
A file in the correct format should look like this:
Site1 CACACTGTGGCCCCTGTGCCCAGCCCTGGGCTCTCTGTACATGAAGCAAC CCCTGTGCCCAGCCC NA NA
Site2 GTCCTGGTTTTTGGTTTGGGAAATATAGTCATC NA GTCCTGGTTTTTGGTTTAAAAAAATATAGTCATC NA
Site 3 TTTCTGGTTTTTGGTTTGGGAAATATAGTCATC NA NA GGAAATATA
The user can easily create this file with any text editor or with spreadsheet software like Excel (Microsoft), Numbers (Apple) or Sheets (Google Docs) and then save it as tab delimited file.
Example:
Using Bioconda:
CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -f AMPLICONS_FILE.txt --name ONLY_AMPLICONS_SRR1046762 --gene_annotations gencode_v19.gz
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -f AMPLICONS_FILE.txt --name ONLY_AMPLICONS_SRR1046762 --gene_annotations gencode_v19.gz
The output of CRISPRessoPooled Amplicons mode consists of:
-
REPORT_READS_ALIGNED_TO_AMPLICONS.txt: this file contains the same information provided in the input description file, plus some additional columns:
a. Demultiplexed_fastq.gz_filename: name of the files containing the raw reads for each amplicon.
b. n_reads: number of reads recovered for each amplicon.
-
A set of fastq.gz files, one for each amplicon.
-
A set of folders, one for each amplicon, containing a full CRISPResso report.
-
SAMPLES_QUANTIFICATION_SUMMARY.txt: this file contains a summary of the quantification and the alignment statistics for each region analyzed (read counts and percentages for the various classes: Unmodified, NHEJ, point mutations, and HDR).
-
CRISPRessoPooled_RUNNING_LOG.txt: execution log and messages for the external utilities called.
Genome mode: In this mode the tool aligns each read to the best location in the genome. Then potential amplicons are discovered looking for regions with enough reads (the default setting is to have at least 1000 reads, but the parameter can be adjusted with the option --min_reads_to_use_region). If a gene annotation file from UCSC is provided, the tool also reports the overlapping gene/s to the region. In this way it is possible to check if the amplified regions map to expected genomic locations and/or also to pseudogenes or other problematic regions. Finally CRISPResso is run in each region discovered.
To run the tool in this mode the user must provide:
-
Paired-end reads (two files) or single-end reads (single file) in FASTQ format (fastq.gz files are also accepted)
-
The full path of the reference genome in bowtie2 format (e.g. /genomes/human_hg19/hg19). Instructions on how to build a custom index or precomputed index for human and mouse genome assembly can be downloaded from the bowtie2 website: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml.
-
Optionally the full path of a gene annotations file from UCSC. The user can download this file from the UCSC Genome Browser ( http://genome.ucsc.edu/cgi-bin/hgTables?command=start ) selecting as table "knownGene", as output format "all fields from selected table" and as file returned "gzip compressed". (e.g. /genomes/human_hg19/gencode_v19.gz)
Example:
Using Bioconda:
CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -x /GENOMES/hg19/hg19 --name ONLY_GENOME_SRR1046762 --gene_annotations gencode_v19.gz
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -x /GENOMES/hg19/hg19 --name ONLY_GENOME_SRR1046762 --gene_annotations gencode_v19.gz
The output of CRISPRessoPooled Genome mode consists of:
- REPORT_READS_ALIGNED_TO_GENOME_ONLY.txt: this file contains the list of all the regions discovered, one per line with the following information:
-
chr_id: chromosome of the region in the reference genome.
-
bpstart: start coordinate of the region in the reference genome.
-
bpend: end coordinate of the region in the reference genome.
-
fastq_file: location of the fastq.gz file containing the reads mapped to the region.
-
n_reads: number of reads mapped to the region.
-
sequence: the sequence, on the reference genome for the region.
-
MAPPED_REGIONS (folder): this folder contains all the fastq.gz files for the discovered regions.
-
A set of folders with the CRISPResso report on the regions with enough reads.
-
SAMPLES_QUANTIFICATION_SUMMARY.txt: this file contains a summary of the quantification and the alignment statistics for each region analyzed (read counts and percentages for the various classes: Unmodified, NHEJ, point mutations, and HDR).
-
CRISPRessoPooled_RUNNING_LOG.txt: execution log and messages for the external utilities called.
This running mode is particularly useful to check for mapping artifacts or contamination in the library. In an optimal experiment, the list of the regions discovered should contain only the regions for which amplicons were designed.
Mixed mode (Amplicons + Genome): in this mode, the tool first aligns reads to the genome and, as in the Genome mode, discovers aligning regions with reads exceeding a tunable threshold. Next it will align the amplicon sequences to the reference genome and will use only the reads that match both the amplicon locations and the discovered genomic locations, excluding spurious reads coming from other regions, or reads not properly trimmed. Finally CRISPResso is run using each of the surviving regions.
To run the tool in this mode the user must provide:
-
Paired-end reads (two files) or single-end reads (single file) in FASTQ format (fastq.gz files are also accepted)
-
A description file containing the amplicon sequences used to enrich regions in the genome and some additional information (as described in the Amplicons mode section).
-
The reference genome in bowtie2 format (as described in Genome mode section).
-
Optionally the gene annotations from UCSC (as described in Genome mode section).
Example:
Using Bioconda:
CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -f AMPLICONS_FILE.txt -x /GENOMES/hg19/hg19 --name AMPLICONS_AND_GENOME_SRR1046762 --gene_annotations gencode_v19.gz
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoPooled -r1 SRR1046762_1.fastq.gz -r2 SRR1046762_2.fastq.gz -f AMPLICONS_FILE.txt -x /GENOMES/hg19/hg19 --name AMPLICONS_AND_GENOME_SRR1046762 --gene_annotations gencode_v19.gz
The output of CRISPRessoPooled Mixed Amplicons + Genome mode consists of these files:
-
REPORT_READS_ALIGNED_TO_GENOME_AND_AMPLICONS.txt: this file contains the same information provided in the input description file, plus some additional columns:
a. Amplicon_Specific_fastq.gz_filename: name of the file containing the raw reads recovered for the amplicon.
b. n_reads: number of reads recovered for the amplicon.
c. Gene_overlapping: gene/s overlapping the amplicon region.
d. chr_id: chromosome of the amplicon in the reference genome.
e. bpstart: start coordinate of the amplicon in the reference genome.
f. bpend: end coordinate of the amplicon in the reference genome.
g. Reference_Sequence: sequence in the reference genome for the region mapped for the amplicon.
-
MAPPED_REGIONS (folder): this folder contains all the fastq.gz files for the discovered regions.
-
A set of folders with the CRISPResso report on the amplicons with enough reads.
-
SAMPLES_QUANTIFICATION_SUMMARY.txt: this file contains a summary of the quantification and the alignment statistics for each region analyzed (read counts and percentages for the various classes: Unmodified, NHEJ, point mutations, and HDR).
-
CRISPRessoPooled_RUNNING_LOG.txt: execution log and messages for the external utilities called.
The Mixed mode combines the benefits of the two previous running modes. In this mode it is possible to recover in an unbiased way all the genomic regions contained in the library, and hence discover contaminations or mapping artifacts. In addition, by knowing the location of the amplicon with respect to the reference genome, reads not properly trimmed or mapped to pseudogenes or other problematic regions will be automatically discarded, providing the cleanest set of reads to quantify the mutations in the target regions with CRISPResso.
If the focus of the analysis is to obtain the best quantification of editing efficiency for a set of amplicons, we suggest running the tool in the Mixed mode. The Genome mode is instead suggested to check problematic libraries, since a report is generated for each region discovered, even if the region is not mappable to any amplicon (however, his may be time consuming). Finally the Amplicon mode is the fastest, although the least reliable in terms of quantification accuracy.
-f or --amplicons_file: Amplicons description file (default: ''). This file is a tab-delimited text file with up to 14 columns (2 required):
--amplicon_name: an identifier for the amplicon (must be unique)
--amplicon_seq: amplicon sequence used in the experiment
--guide_seq (OPTIONAL): sgRNA sequence used for this amplicon without the PAM sequence. Multiple guides can be given separated by commas and not spaces.
--expected_hdr_amplicon_seq (OPTIONAL): expected amplicon sequence in case of HDR.
--coding_seq (OPTIONAL): Subsequence(s) of the amplicon corresponding to coding sequences. If more than one separate them by commas and not spaces.
--prime_editing_pegRNA_spacer_seq (OPTIONAL): pegRNA spacer sgRNA sequence used in prime editing. The spacer should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the given sequence.
--prime_editing_nicking_guide_seq (OPTIONAL): Nicking sgRNA sequence used in prime editing. The sgRNA should not include the PAM sequence. The sequence should be given in the RNA 5'->3' order, so for Cas9, the PAM would be on the right side of the sequence.
--prime_editing_pegRNA_extension_seq (OPTIONAL): Extension sequence used in prime editing. The sequence should be given in the RNA 5'->3' order, such that the sequence starts with the RT template including the edit, followed by the Primer-binding site (PBS).
--prime_editing_pegRNA_scaffold_seq (OPTIONAL): If given, reads containing any of this scaffold sequence before extension sequence (provided by --prime_editing_extension_seq) will be classified as 'Scaffold-incorporated'. The sequence should be given in the 5'->3' order such that the RT template directly follows this sequence. A common value ends with 'GGCACCGAGUCGGUGC'.
--prime_editing_pegRNA_scaffold_min_match_length (OPTIONAL): Minimum number of bases matching scaffold sequence for the read to be counted as 'Scaffold-incorporated'. If the scaffold sequence matches the reference sequence at the incorporation site, the minimum number of bases to match will be minimally increased (beyond this parameter) to disambiguate between prime-edited and scaffold-incorporated sequences.
--prime_editing_override_prime_edited_ref_seq (OPTIONAL): If given, this sequence will be used as the prime-edited reference sequence. This may be useful if the prime-edited reference sequence has large indels or the algorithm cannot otherwise infer the correct reference sequence.
--quantification_window_coordinates (OPTIONAL): Bp positions in the amplicon sequence specifying the quantification window. This parameter overrides values of the "--quantification_window_center", "-- cleavage_offset", "--window_around_sgrna" or "-- window_around_sgrna" values. Any indels/substitutions outside this window are excluded. Indexes are 0-based, meaning that the first nucleotide is position 0. Ranges are separated by the dash sign like "start-stop", and multiple ranges can be separated by the underscore (_). A value of 0 disables this filter. (can be comma-separated list of values, corresponding to amplicon sequences given in --amplicon_seq e.g. 5-10,5-10_20-30 would specify the 5th-10th bp in the first reference and the 5th-10th and 20th-30th bp in the second reference) (default: None)
--quantification_window_size (OPTIONAL): Defines the size (in bp) of the quantification window extending from the position specified by the "--cleavage_offset" or "--quantification_window_center" parameter in relation to the provided guide RNA sequence(s) (--sgRNA). Mutations within this number of bp from the quantification window center are used in classifying reads as modified or unmodified. A value of 0 disables this window and indels in the entire amplicon are considered. Default is 1, 1bp on each side of the cleavage position for a total length of 2bp.
--quantification_window_center (OPTIONAL): Center of quantification window to use within respect to the 3' end of the provided sgRNA sequence. Remember that the sgRNA sequence must be entered without the PAM. For cleaving nucleases, this is the predicted cleavage position. The default is -3 and is suitable for the Cas9 system. For alternate nucleases, other cleavage offsets may be appropriate, for example, if using Cpf1 this parameter would be set to 1. For base editors, this could be set to -17.
--gene_annotations: Gene Annotation Table from UCSC Genome Browser Tables http://genome.ucsc.edu/cgi-bin/hgTables?command=start, please select as table "knownGene", as output format "all fields from selected table" and as file returned "gzip compressed". (default: '')
-x or --bowtie2_index: Basename of Bowtie2 index for the reference genome. (default: '')
--bowtie2_options_string: Override options for the Bowtie2 alignment command. By default, this is " --end-to-end -N 0 --np 0 -mp 3,2 --score-min L,-5,-3(1-H)" where H is the default homology score. (default: ' --end-to-end -N 0 --np 0 -mp 3,2 --score-min L,-5,-3(1-H)')
--use_legacy_bowtie2_options_string: Use legacy (more stringent) Bowtie2 alignment parameters: " -k 1 --end-to-end -N 0 --np 0 ". (default: False)
--min_reads_to_use_region: Minimum number of reads that align to a region to perform the CRISPResso analysis. (default: 1000)
--skip_failed: Continue with pooled analysis even if one sample fails. (default: False)
--skip_reporting_problematic_regions: Skip reporting of problematic regions. By default, when both amplicons (-f) and genome (-x) are provided, problematic reads that align to the genome but to positions other than where the amplicons align are reported as problematic. (default: False)
--compile_postrun_references: If set, a file will be produced which compiles the reference sequences of frequent amplicons. (default: False)
--compile_postrun_reference_allele_cutoff: Only alleles with at least this percentage frequency in the population will be reported in the postrun analysis. This parameter is given as a percent, so 30 is 30%. (default: 30)
--alternate_alleles: Path to tab-separated file with alternate allele sequences for pooled experiments. This file has the columns "region_name","reference_seqs", and "reference_names" and gives the reference sequences of alternate alleles that will be passed to CRISPResso for each individual region for allelic analysis. Multiple reference alleles and reference names for a given region name are separated by commas (no spaces). (default: '')
--limit_open_files_for_demux: If set, only one file will be opened during demultiplexing of read alignment locations. This will be slightly slower as the reads must be sorted, but may be necessary if the number of amplicons is greater than the number of files that can be opened due to OS constraints. (default: False)
CRISPRessoWGS is a utility for the analysis of genome editing experiment from whole genome sequencing (WGS) data. CRISPRessoWGS allows exploring any region of the genome to quantify targeted editing or potentially off-target effects. The intended use case for CRISPRessoWGS is the analysis of targeted regions, and WGS reads from those regions will be realigned using CRISPResso's alignment aligorithm for more accurate genome editing quantification. To scan the entire genome for mutations VarScan or MuTect are more suitable, and identified regions can be analyzed and visualized using CRISPRessoWGS.
To run CRISPRessoWGS you must provide:
-
A genome aligned BAM file. To align reads from a WGS experiment to the genome there are many options available, we suggest using either Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/) or BWA (http://bio-bwa.sourceforge.net/).
-
A FASTA file containing the reference sequence used to align the reads and create the BAM file (the reference files for the most common organism can be download from UCSC: http://hgdownload.soe.ucsc.edu/downloads.html. Download and uncompress only the file ending with .fa.gz, for example for the last version of the human genome download and uncompress the file hg38.fa.gz)
-
Descriptions file containing the coordinates of the regions to analyze and some additional information. In particular, this file is a tab delimited text file with up to 7 columns (4 required):
-
chr_id: chromosome of the region in the reference genome.
-
bpstart: start coordinate of the region in the reference genome.
-
bpend: end coordinate of the region in the reference genome.
-
REGION_NAME: an identifier for the region (must be unique).
-
sgRNA_SEQUENCE (OPTIONAL): sgRNA sequence used for this genomic segment without the PAM sequence. If not available, enter NA.
-
EXPECTED_SEGMENT_AFTER_HDR (OPTIONAL): expected genomic segment sequence in case of HDR. If more than one, separate by commas and not spaces. If not available, enter NA.
-
CODING_SEQUENCE (OPTIONAL): Subsequence(s) of the genomic segment corresponding to coding sequences. If more than one, separate by commas and not spaces. If not available, enter NA.
-
A file in the correct format should look like this (column entries must be separated by tabs):
chr1 65118211 65118261 R1 CTACAGAGCCCCAGTCCTGG NA NA
chr6 51002798 51002820 R2 NA NA NA
Note: no column titles should be entered. As you may have noticed this file is just a BED file with extra columns. For this reason a normal BED file with 4 columns, is also accepted by this utility.
- Optionally the full path of a gene annotations file from UCSC. You can download the this file from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTables?command=start) selecting as table "knownGene", as output format "all fields from selected table" and as file returned "gzip compressed". (something like: /genomes/human_hg19/gencode_v19.gz)
Example:
Using Bioconda:
CRISPRessoWGS -b WGS/50/50_sorted_rmdup_fixed_groups.bam -f WGS_TEST.txt -r /GENOMES/mm9/mm9.fa --gene_annotations ensemble_mm9.txt.gz --name CRISPR_WGS_SRR1542350
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoWGS -b WGS/50/50_sorted_rmdup_fixed_groups.bam -f WGS_TEST.txt -r /GENOMES/mm9/mm9.fa --gene_annotations ensemble_mm9.txt.gz --name CRISPR_WGS_SRR1542350
The output from these files will consist of:
-
REPORT_READS_ALIGNED_TO_SELECTED_REGIONS_WGS.txt: this file contains the same information provided in the input description file, plus some additional columns:
a. sequence: sequence in the reference genome for the region specified.
b. gene_overlapping: gene/s overlapping the region specified.
c. n_reads: number of reads recovered for the region.
d. bam_file_with_reads_in_region: file containing only the subset of the reads that overlap, also partially, with the region. This file is indexed and can be easily loaded for example on IGV for visualization of single reads or for the comparison of two conditions. For example, in the figure below (fig X) we show reads mapped to a region inside the coding sequence of the gene Crygc subjected to NHEJ (CRISPR_WGS_SRR1542350) vs reads from a control experiment (CONTROL_WGS_SRR1542349).
e. fastq.gz_file_trimmed_reads_in_region: file containing only the subset of reads fully covering the specified regions, and trimmed to match the sequence in that region. These reads are used for the subsequent analysis with CRISPResso.
-
ANALYZED_REGIONS (folder): this folder contains all the BAM and FASTQ files, one for each region analyzed.
-
A set of folders with the CRISPResso report on the regions provided in input with enough reads (the default setting is to have at least 10 reads, but the parameter can be adjusted with the option
--min_reads_to_use_region).
-
CRISPRessoPooled_RUNNING_LOG.txt: execution log and messages for the external utilities called.
This utility is particular useful to investigate and quantify mutation frequency in a list of potential target or off-target sites, coming for example from prediction tools, or from other orthogonal assays.
-b or --bam_file: WGS aligned bam file. (default: 'bam filename')
-f or --region_file: Regions description file. A BED format file containing the regions to analyze, one per line. The REQUIRED columns are: chr_id(chromosome name), bpstart(start position), bpend(end position), the optional columns are: name (an unique indentifier for the region), guide_seq, expected_hdr_amplicon_seq,coding_seq, see CRISPResso help for more details on these last 3 parameters).
-r or --reference_file: A FASTA format reference file (for example hg19.fa for the human genome). (default: '')
--min_reads_to_use_region: Minimum number of reads that align to a region to perform the CRISPResso analysis. (default: 10)
--gene_annotations: Gene Annotation Table from UCSC Genome Browser Tables http://genome.ucsc.edu/cgi-bin/hgTables?command=start, please select as table "knownGene", as output format "all fields from selected table" and as file returned "gzip compressed". (default: '')
--crispresso_command: CRISPResso command to call. (default: 'CRISPResso')
CRISPRessoCompare is a utility for the comparison of a pair of CRISPResso analyses. CRISPRessoCompare produces a summary of differences between two conditions, for example a CRISPR treated and an untreated control sample (see figure below). Informative plots are generated showing the differences in editing rates and localization within the reference amplicon,
To run CRISPRessoCompare you must provide:
- Two output folders generated with CRISPResso using the same reference amplicon and settings but on different datasets.
- Optionally a name for each condition to use for the plots, and the name of the output folder
Example:
Using Bioconda:
CRISPRessoCompare -n1 "VEGFA CRISPR" -n2 "VEGFA CONTROL" -n VEGFA_Site_1_SRR10467_VS_SRR1046787 CRISPResso_on_VEGFA_Site_1_SRR1046762/ CRISPResso_on_VEGFA_Site_1_SRR1046787/
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoCompare -n1 "VEGFA CRISPR" -n2 "VEGFA CONTROL" -n VEGFA_Site_1_SRR10467_VS_SRR1046787 CRISPResso_on_VEGFA_Site_1_SRR1046762/ CRISPResso_on_VEGFA_Site_1_SRR1046787/
The output will consist of:
- Comparison_Efficiency.pdf: a figure containing a comparison of the edit frequencies for each category (NHEJ, MIXED NHEJ-HDR and HDR) and as well the net effect subtracting the second sample (second folder in the command line) provided in the analysis from the first sample (first folder in the command line).
- Comparison_Combined_Insertion_Deletion_Substitution_Locations.pdf: a figure showing the average profile for the mutations for the two samples in the same scale and their difference with the same convention used in the previous figure (first sample – second sample).
- CRISPRessoCompare_significant_base_counts.txt: a text file reporting the number of bases for each amplicon and in the quantification window for each amplicon that were significantly enriched for Insertions, Deletions, and Substitutions, as well as All Modifications (Fisher's exact test, Bonferonni corrected p-values).
- CRISPRessoCompare_RUNNING_LOG.txt: detailed execution log.
crispresso_output_folder_1: First output folder with CRISPResso analysis (Required) crispresso_output_folder_2: Second output folder with CRISPResso analysis (Required)
-n or --name: Output name. (default:'') -n1 or --sample_1_name: Sample 1 name -n2 or --sample_2_name: Sample 2 name -o or --output_folder: Output folder name --reported_qvalue_cutoff: Q-value cutoff for signifance in tests for differential editing. Each base position is tested (for insertions, deletions, substitutions, and all modifications) using Fisher's exact test, followed by Bonferonni correction. The number of bases with a significance below this threshold in the quantification window are counted and reported in the output summary. (default:0.05) --min_frequency_alleles_around_cut_to_plot: Minimum %% reads required to report an allele in the alleles table plot. (default:0.2) --max_rows_alleles_around_cut_to_plot: Maximum number of rows to report in the alleles table plot. (default:50) --suppress_report: Suppress output report. (default:False) --place_report_in_output_folder: If true, report will be written inside the CRISPResso output folder. By default, the report will be written one directory up from the report output. (default:False)
CRISPRessoPooledWGSCompare is an extension of the CRIPRessoCompare utility allowing the user to run and summarize multiple CRISPRessoCompare analyses where several regions are analyzed in two different conditions, as in the case of the CRISPRessoPooled or CRISPRessoWGS utilities.
To run CRISPRessoPooledWGSCompare you must provide:
- Two output folders generated with CRISPRessoPooled or CRISPRessoWGS using the same reference amplicon and settings but on different datasets.
- Optionally a name for each condition to use for the plots, and the name of the output folder
Example:
Using Bioconda:
CRISPRessoPooledWGSCompare CRISPRessoPooled_on_AMPLICONS_AND_GENOME_SRR1046762/ CRISPRessoPooled_on_AMPLICONS_AND_GENOME_SRR1046787/ -n1 SRR1046762 -n2 SRR1046787 -n AMPLICONS_AND_GENOME_SRR1046762_VS_SRR1046787
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoPooledWGSCompare CRISPRessoPooled_on_AMPLICONS_AND_GENOME_SRR1046762/ CRISPRessoPooled_on_AMPLICONS_AND_GENOME_SRR1046787/ -n1 SRR1046762 -n2 SRR1046787 -n AMPLICONS_AND_GENOME_SRR1046762_VS_SRR1046787
The output from these files will consist of:
- COMPARISON_SAMPLES_QUANTIFICATION_SUMMARIES.txt: this file contains a summary of the quantification for each of the two conditions for each region and their difference (read counts and percentages for the various classes: Unmodified, NHEJ, MIXED NHEJ-HDR and HDR).
- A set of folders with CRISPRessoCompare reports on the common regions with enough reads in both conditions.
- CRISPRessoPooledWGSCompare_significant_base_count_summary.txt: a text file summarizing for each sample and amplicon in both conditions the number of bases for each amplicon and in the quantification window for each amplicon that were significantly enriched for Insertions, Deletions, and Substitutions, as well as All Modifications (Fisher's exact test, Bonferonni corrected p-values).
- CRISPRessoPooledWGSCompare_RUNNING_LOG.txt: detailed execution log.
crispresso_pooled_wgs_output_folder_1: First output folder with CRISPRessoPooled or CRISPRessoWGS analysis (Required) crispresso_pooled_wgs_output_folder_2: Second output folder with CRISPRessoPooled or CRISPRessoWGS analysis (Required)
-p or --n_processes: Number of processes to use for this analysis. Can be set to 'max'. -n or --name: Output name. (default:'') -n1 or --sample_1_name: Sample 1 name -n2 or --sample_2_name: Sample 2 name -o or --output_folder: Output folder name --reported_qvalue_cutoff: Q-value cutoff for signifance in tests for differential editing. Each base position is tested (for insertions, deletions, substitutions, and all modifications) using Fisher's exact test, followed by Bonferonni correction. The number of bases with a significance below this threshold in the quantification window are counted and reported in the output summary. (default:0.05) --min_frequency_alleles_around_cut_to_plot: Minimum %% reads required to report an allele in the alleles table plot. (default:0.2) --max_rows_alleles_around_cut_to_plot: Maximum number of rows to report in the alleles table plot. (default:50) --suppress_report: Suppress output report. (default:False) --place_report_in_output_folder: If true, report will be written inside the CRISPResso output folder. By default, the report will be written one directory up from the report output. (default:False)
CRISPRessoAggregate is a utility to combine the analysis of several CRISPResso runs. The analyses are summarized and editing rates are optionally visualized in a summary report.
CRISPRessoAggregate has the following parameters:
--name: Output name of the report (required)
--prefix: Prefix for CRISPResso folders to aggregate (may be specified multiple times)
--suffix: Suffix for CRISPResso folders to aggregate
--min_reads_for_inclusion: Minimum number of reads for a run to be included in the run summary (default: 0)
--place_report_in_output_folder: If true, report will be written inside the CRISPResso output folder. By default, the report will be written one directory up from the report output (default: False)
--suppress_report: Suppress output report (default: False)
--suppress_plots: Suppress output plots (default: False)
To run CRISPRessoCompare you must provide the --name parameter, and CRISPResso folders in the current directory will be summarized. To summarize folders in other locations, provide these locations using the '--prefix' parameter.
Example:
Using Bioconda:
CRISPRessoAggregate --name "VEGFA" --prefix CRISPRessoRuns/VEGFA/
Using Docker:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPRessoAggregate --name "VEGFA" --prefix CRISPRessoRuns/VEGFA/
The output will consist of:
- CRISPResso2Aggregate_report.html: a html file containing links to all aggregated runs.
- CRISPRessoAggregate_amplicon_information.txt: A tab-separated file with a line for each amplicon that was found in any run. The 'Amplicon Name' column shows the unique name for this amplicon sequence. 'Number of sources' shows how many runs the amplicon was found in, and 'Amplicon sources' show which run folders the amplicon was found in, as well as the name of the amplicon in that run.
- CRISPRessoAggregate_mapping_statistics.txt: A tab-separated file showing the number of reads sequenced and mapped for each run.
- CRISPRessoAggregate_quantification_of_editing_frequency.txt: A tab-separated with the number of reads and edits for each run folder. Data from run folders with multiple amplicons show the sum totals for all amplicons.
- CRISPRessoAggregate_quantification_of_editing_frequency_by_amplicon.txt: A tab-separated file showing the number of reads and edits for each amplicon for each run folder. Data from run folders with multiple amplicons will appear on multiple lines, with one line per amplicon.
CRISPResso is an open source tool for free use by academics. However, for-profit organizations are required to purchase a license to use CRISPResso. As a part of this license, organizations gain access to the CRISPRessoPro package which supplements CRISPResso with several useful features:
- Interactive and improved plots using D3 and Plotly
- Customizable colors
- Customizable warnings based on potential issues in results (guardrails)
To add CRISPRessoPro to CRISPResso contact Edilytics - [email protected]
If CRISPRessoPro is installed, by default reports will include interactive plots. To use matplotlib for figures add the --use_matplotlib argument.
If CRISPRessoPro is installed, by default the colors and guardrails will remain the same as CRISPResso. To alter this, use the --config_file argument and a filepath to a .json file with the following format:
{
"colors": {
"Substitution": "#0000FF",
"Insertion": "#008000",
"Deletion": "#FF0000",
"A": "#7FC97F",
"T": "#BEAED4",
"C": "#FDC086",
"G": "#FFFF99",
"N": "#C8C8C8",
"-": "#1E1E1E"
},
"guardrails": {
"min_total_reads": 10000,
"aligned_cutoff": 0.9,
"alternate_alignment": 0.3,
"min_ratio_of_mods_in_to_out": 0.01,
"modifications_at_ends": 0.01,
"outside_window_max_sub_rate": 0.002,
"max_rate_of_subs": 0.3,
"guide_len": 19,
"amplicon_len": 50,
"amplicon_to_read_length": 1.5
}
}Above are the default values as an example, change the values as desired to any color or guardrail specification.