A Fiji plugin for loading and saving image data to N5 containers. Supports HDF5, Zarr, Amazon S3, and Google cloud storage.
Open N5 datasets from Fiji with File > Import > N5.
Quickly open a dataset by pasting the full path to the dataset and press OK.
For example, try gs://example_multi-n5_bucket/mitosis.n5/raw to open the sample mitosis image from Google
cloud storage.
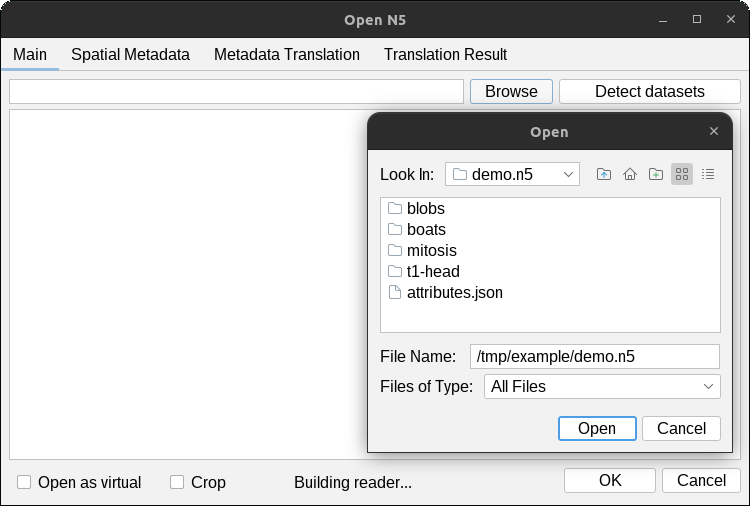
Click the Browse button to select a folder on your filesystem.
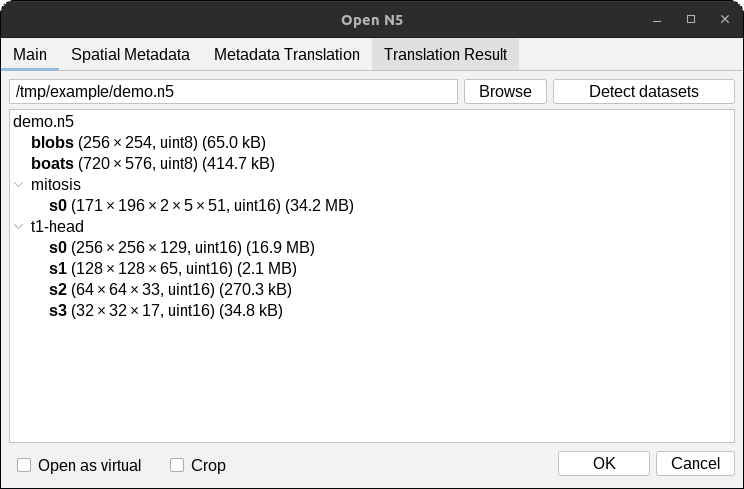
The detected datasets will be displayed in the dialog. Select (highlight) the datasets you would like to open
and press OK. In the example below, we will open the datasets /blobs, and /t1-head/c0/s0.
Check the Open as virtual box to open the n5 dataset as a virtual stack in ImageJ.
This enable the opening and viewing of image data that do not fit in RAM. Image slices are loaded on-the-fly, so
navigation will be slow when parts of the images are loaded.
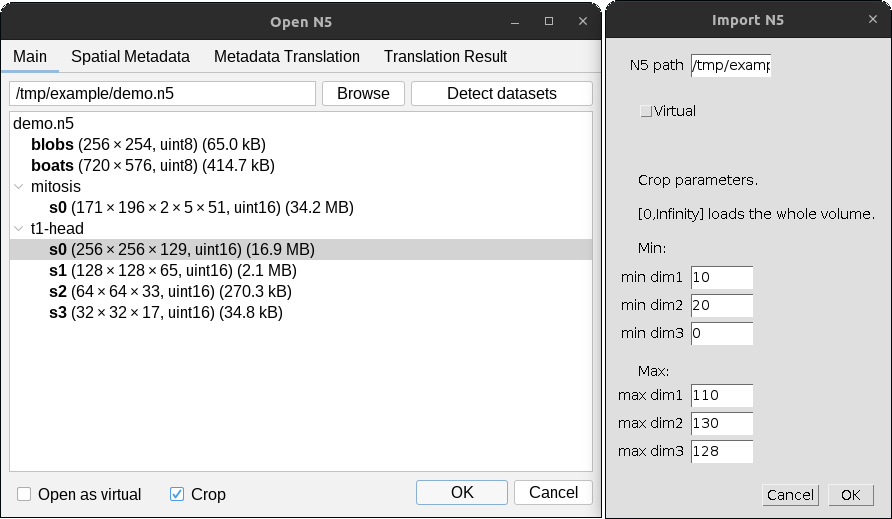
Subsets of images can be opened by checking the Crop box in the dialog, then pressing OK.
A separate dialog will appear for each selected dataset as shown below.
Give the min and max values for the field-of-view to open in pixel / voxel units a particular
subset. The opened interval includes both min and max values, so the image will be of size max - min + 1 along
each dimension. In the example shown above, the resulting image will be of size 101 x 111 x 2 x 51.
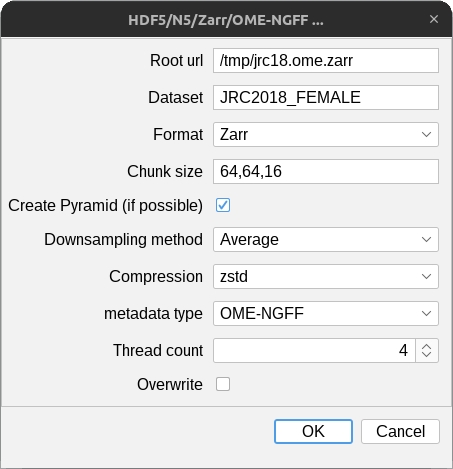
Save images opened in Fiji as N5 datasets with File > Save As > Export N5.
Parameters
N5Root- the root location of the n5 (see also Container types)Dataset- the name of the dataset.Chunk size- chunk/block size as comma-separated list.- ImageJ's axis order is X,Y,C,Z,T. The chunk size must be specified in this order. You must skip any axis whose size is
1, e.g. a 2D time-series without channels may have a chunk size of1024,1024,1(X,Y,T). - You may provide fewer values than the data dimension. In that case, the list will be expanded to necessary size with the last value, for example
64, will expand to64,64,64for 3D data.
- ImageJ's axis order is X,Y,C,Z,T. The chunk size must be specified in this order. You must skip any axis whose size is
Create Pyramid- If checked, a multiscale pyramid will be created (if possible). See below for details.Downsampling method- The downsampling method to be used if a multiscale pyramid can be created. See below for details.Compression- The compression method to be used for chucnks / blocks.metadata type- style and type of metadata to store (see also Metadata)thread count- number of threads used for parallel writing (see also Cloud writing benchmarks)Overwrite- If checked, existing data may be deleted and overwritten without warning.
The export plugin infers container type from the file/directory path or url given as the n5 root:
- Filesystem N5
- Specify a directory ending in
.n5 - example
/path/to/my/data.n5
- Specify a directory ending in
- Zarr
- Specify a directory ending in
.zarr - example
/Users/user/Documents/sample.zarr
- Specify a directory ending in
- HDF5
- Specify a file ending in
.h5,.hdf5, or.hdf - example
C:\user\docs\example.h5
- Specify a file ending in
- Amazon S3
- Specify one of two url styles:
s3://bucket-name/path/to/root.n5https://bucket-name.s3.amazonaws.com/path/to/root.n5
- Google cloud storage (one of two url styles)
- Specify one of two url styles:
gs://bucket-name/path/inside/bucket/root.n5https://bucket-name.s3.amazonaws.com/path/to/root.n5
The number of scale levels is determined by the image size and specified block size. The exporter will downsample an image only if the block size is striclty smaller than the image size in every dimension.
If image size is 100 x 100 x 100 and the block size is 100 x 100 x 100
will write one scale level because the whole image fits into one block
at the first scale level.
If image size is 100 x 100 x 100 and the block size is 64 x 64 x 64
will write two scale levels: The first scale level will have a 2 x 2 x 2 grid
of blocks.
If image size is 100 x 100 x 32 and the block size is 64 x 64 x 64
will write one scale level because the third dimension of the image is
smaller than the third dimension of the block.
The N5 exporter always downsamples images by factors of two in all dimensions. There are two downsampling methods:
N5 will take even-indexed samples and discard odd-indexed samples along each dimension.
N5 will average adjacent samples along each dimension. This results in a "half-pixel" shift, which will be reflected in the metadata.
If the overwrite option is not selected in the dialog, the exporter will determine if the
write operation would overwrite or invalidate existing data. If so, it prompts the user with a
warning dialog, asking if data should be overwritten.
Data could be overwritten if:
- the path where data will be written already exists and contains data.
- a parent of the path where data will be written already exists and contains data.
- here, the newly written data would be inaccessible, because data arrays must be leafs of the hierarchy tree.
If the overwrite option is selected in the initial dialog, the user will not be prompted, but
data will be overwritten if needed as explained below.
A dataset exists at /image. User attempts to write data into /image. This warns the user
about overwriting because an array already exists at that location.
A dataset exists at /data/s0. User attempts to write data into /data using N5Viewer metadata.
This warns the user about overwriting because when writing N5Viewer metadata, the plugin will
write the full resolution level of the multiscale pyramid at location /data/s0, but an array
already exists at that location.
If the user decides to overwite data the N5 exporter will completely (array data and metadata) remove any groups that cause conflicts before writing the new data.
- If a dataset already exists at a path where new data will be written, then all data at that path will be removed.
- If a dataset already exists at a parent path where new data will be written, then all data at that parent path will be removed.
A dataset exists at /image. User attempts to write data into /image/channel0. This warns
the user about overwriting because the newly written data would be a child path of existing
data, and therefore be invalid. If the user decides to overwrite, all data at /image will be
removed before writing the new data to /image/channel0.
This plugin supports three types of image metadata:
- ImageJ-style metadata
- N5-viewer metadata
- COSEM metadata
- Custom metadata. Read details here
The metadata style for exported N5 datasets is customizable, more detail coming soon.
ImageJ convenience layer for N5
Build into your Fiji installation:
mvn -Dscijava.app.directory=/home/saalfelds/packages/Fiji.app -Ddelete.other.versions=true clean installThen, in Fiji's Script Interpreter (Plugins > Scripting > Script Interpreter), load an N5 dataset into an ImagePlus:
import org.janelia.saalfeldlab.n5.*;
import org.janelia.saalfeldlab.n5.ij.*;
imp = N5IJUtils.load(new N5FSReader("/home/saalfelds/example.n5"), "/volumes/raw");or save an ImagePlus into an N5 dataset:
import ij.IJ;
import org.janelia.saalfeldlab.n5.*;
import org.janelia.saalfeldlab.n5.ij.*;
N5IJUtils.save(
IJ.getImage(),
new N5FSWriter("/home/saalfelds/example.n5"),
"/volumes/raw",
new int[] {128, 128, 64},
new GzipCompression()
);Save an image stored locally to cloud storage (using four threads):
final ImagePlus imp = IJ.openImage( "/path/to/some.tif" );
final ExecutorService exec = Executors.newFixedThreadPool( 4 );
N5IJUtils.save( imp,
new N5Factory().openWriter( "s3://myBucket/myContainer.n5" ),
"/myDataset",
new int[]{64, 64, 64},
new GzipCompression(),
exec );
exec.shutdown();See also scripts demonstrating
- how to read and write imglib2 images with the methods in
N5Utils - how to read and write ImageJ images with the methods in
N5IJUtils
-
This plugin supports images of up to 5 dimensions, and the datatypes supported by ImageJ (
uint8,uint16,float32) For higher dimensions and other datatypes, we recommend n5-imglib2. -
This plugin supports only the datatypes supported by ImageJ, namely uint8, uint16, and float32. For other datatypes, use n5-imglib2.
Below are benchmarks for writing images of various sizes, block sizes, and with increasing amount of parallelism.
Time in seconds to write the image data. Increased parallelism speeds up writing substantially when the total number of blocks is high.
| Image size | Block size | 1 thread | 2 threads | 4 threads | 8 threads | 16 threads |
|---|---|---|---|---|---|---|
| 64x64x64 | 32x32x32 | 0.98 | 0.60 | 0.45 | 0.50 | 0.51 |
| 128x128x128 | 32x32x32 | 4.72 | 2.64 | 1.62 | 1.00 | |
| 256x256x256 | 32x32x32 | 37.09 | 19.11 | 9.09 | 5.20 | 3.2 |
| 256x256x256 | 64x64x64 | 10.56 | 5.04 | 3.23 | 2.17 | 1.86 |
| 512x512x512 | 32x32x32 | 279.28 | 156.89 | 74.72 | 37.15 | 19.77 |
| 512x512x512 | 64x64x64 | 76.63 | 38.16 | 19.86 | 10.16 | 6.14 |
| 512x512x512 | 128x128x128 | 27.16 | 14.32 | 8.01 | 4.70 | 3.31 |
| 1024x1024x1024 | 32x32x32 | 2014.73 | 980.66 | 483.04 | 249.83 | 122.36 |
| 1024x1024x1024 | 64x64x64 | 579.46 | 289.53 | 149.98 | 75.85 | 38.18 |
| 1024x1024x1024 | 128x128x128 | 203.47 | 107.23 | 55.11 | 27.41 | 15.33 |